Phylogenetic reconstruction of ancestral character states for gene expression and mRNA splicing data
- PMID: 15921519
- PMCID: PMC1166541
- DOI: 10.1186/1471-2105-6-127
Phylogenetic reconstruction of ancestral character states for gene expression and mRNA splicing data
Abstract
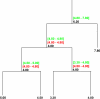
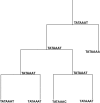
Background: As genomes evolve after speciation, gene content, coding sequence, gene expression, and splicing all diverge with time from ancestors with close relatives. A minimum evolution general method for continuous character analysis in a phylogenetic perspective is presented that allows for reconstruction of ancestral character states and for measuring along branch evolution.
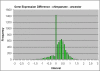
Results: A software package for reconstruction of continuous character traits, like relative gene expression levels or alternative splice site usage data is presented and is available for download at http://www.rossnes.org/phyrex. This program was applied to a primate gene expression dataset to detect transcription factor binding sites that have undergone substitution, potentially having driven lineage-specific differences in gene expression.
Conclusion: Systematic analysis of lineage-specific evolution is becoming the cornerstone of comparative genomics. New methods, like phyrex, extend the capabilities of comparative genomics by tracing the evolution of additional biomolecular processes.
Figures



Similar articles
-
EuSplice: a unified resource for the analysis of splice signals and alternative splicing in eukaryotic genes.Bioinformatics. 2007 Jul 15;23(14):1815-23. doi: 10.1093/bioinformatics/btm084. Epub 2007 Mar 7. Bioinformatics. 2007. PMID: 17344236
-
Characterization of 954 bovine full-CDS cDNA sequences.BMC Genomics. 2005 Nov 23;6:166. doi: 10.1186/1471-2164-6-166. BMC Genomics. 2005. PMID: 16305752 Free PMC article.
-
Protein encoding genes in an ancient plant: analysis of codon usage, retained genes and splice sites in a moss, Physcomitrella patens.BMC Genomics. 2005 Mar 22;6:43. doi: 10.1186/1471-2164-6-43. BMC Genomics. 2005. PMID: 15784153 Free PMC article.
-
Moving primate genomics beyond the chimpanzee genome.Trends Genet. 2005 Sep;21(9):511-7. doi: 10.1016/j.tig.2005.06.012. Trends Genet. 2005. PMID: 16009448 Review.
-
Comparative primate genomics: the year of the chimpanzee.Curr Opin Genet Dev. 2004 Dec;14(6):650-6. doi: 10.1016/j.gde.2004.08.007. Curr Opin Genet Dev. 2004. PMID: 15531160 Review.
Cited by
-
Characterization of an environmental strain of Bacillus thuringiensis from a hot spring in Western Himalayas.Curr Microbiol. 2011 Feb;62(2):547-56. doi: 10.1007/s00284-010-9743-x. Epub 2010 Aug 25. Curr Microbiol. 2011. PMID: 20737272
-
The Adaptive Evolution Database (TAED): a phylogeny based tool for comparative genomics.Nucleic Acids Res. 2005 Jan 1;33(Database issue):D495-7. doi: 10.1093/nar/gki090. Nucleic Acids Res. 2005. PMID: 15608245 Free PMC article.
-
Measuring gene expression divergence: the distance to keep.Biol Direct. 2010 Aug 6;5:51. doi: 10.1186/1745-6150-5-51. Biol Direct. 2010. PMID: 20691088 Free PMC article.
-
Datasets for evolutionary comparative genomics.Genome Biol. 2005;6(8):117. doi: 10.1186/gb-2005-6-8-117. Epub 2005 Jul 28. Genome Biol. 2005. PMID: 16086856 Free PMC article.
-
Comparative analysis indicates regulatory neofunctionalization of yeast duplicates.Genome Biol. 2007;8(4):R50. doi: 10.1186/gb-2007-8-4-r50. Genome Biol. 2007. PMID: 17411427 Free PMC article.
References
-
- Fitch WM. Toward defining the course of evolution: Minimal change for a specific tree topology. Syst Zool. 1971;19:99–113.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

