The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis
- PMID: 15923334
- PMCID: PMC1150422
- DOI: 10.1104/pp.104.058032
The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis
Abstract
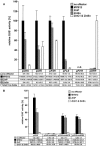
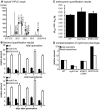
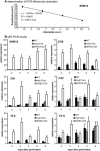
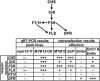
Comprehensive functional data on plant R2R3-MYB transcription factors is still scarce compared to the manifold of their occurrence. Here, we identified the Arabidopsis (Arabidopsis thaliana) R2R3-MYB transcription factor MYB12 as a flavonol-specific activator of flavonoid biosynthesis. Transient expression in Arabidopsis protoplasts revealed a high degree of functional similarity between MYB12 and the structurally closely related factor P from maize (Zea mays). Both displayed similar target gene specificity, and both activated target gene promoters only in the presence of a functional MYB recognition element. The genes encoding the flavonoid biosynthesis enzymes chalcone synthase, chalcone flavanone isomerase, flavanone 3-hydroxylase, and flavonol synthase were identified as target genes. Hence, our observations further add to the general notion of a close relationship between structure and function of R2R3-MYB factors. High-performance liquid chromatography analyses of myb12 mutant plants and MYB12 overexpression plants demonstrate a tight linkage between the expression level of functional MYB12 and the flavonol content of young seedlings. Quantitative real time reverse transcription-PCR using these mutant plants showed MYB12 to be a transcriptional regulator of CHALCONE SYNTHASE and FLAVONOL SYNTHASE in planta, the gene products of which are indispensable for the biosynthesis of flavonols.
Figures




References
-
- Andersson KB, Berge T, Matre V, Gabrielsen OS (1999) Sequence selectivity of c-Myb in vivo. Resolution of a DNA target specificity paradox. J Biol Chem 274: 21986–21994 - PubMed
-
- Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L (2004) TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J 39: 366–380 - PubMed
-
- Bechtold N, Ellis J, Pelletier G (1993) In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Paris Life Sci 316: 1194–1199
-
- Becker D, Kemper E, Schell J, Masterson R (1992) New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol Biol 20: 1195–1197 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

