Constitutively high expression of the histidine biosynthetic pathway contributes to nickel tolerance in hyperaccumulator plants
- PMID: 15923352
- PMCID: PMC1167554
- DOI: 10.1105/tpc.104.030577
Constitutively high expression of the histidine biosynthetic pathway contributes to nickel tolerance in hyperaccumulator plants
Abstract
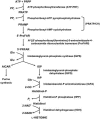
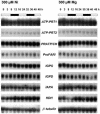
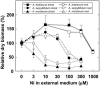
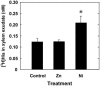
Plants that hyperaccumulate Ni exhibit an exceptional degree of Ni tolerance and the ability to translocate Ni in large amounts from root to shoot. In hyperaccumulator plants in the genus Alyssum, free His is an important Ni binding ligand that increases in the xylem proportionately to root Ni uptake. To determine the molecular basis of the His response and its contribution to Ni tolerance, transcripts representing seven of the eight enzymes involved in His biosynthesis were investigated in the hyperaccumulator species Alyssum lesbiacum by RNA gel blot analysis. None of the transcripts changed in abundance in either root or shoot tissue when plants were exposed to Ni, but transcript levels were constitutively higher in A. lesbiacum than in the congeneric nonaccumulator A. montanum, especially for the first enzyme in the biosynthetic pathway, ATP-phosphoribosyltransferase (ATP-PRT). Comparison with the weak hyperaccumulator A. serpyllifolium revealed a close correlation between Ni tolerance, root His concentration, and ATP-PRT transcript abundance. Overexpression of an A. lesbiacum ATP-PRT cDNA in transgenic Arabidopsis thaliana increased the pool of free His up to 15-fold in shoot tissue, without affecting the concentration of any other amino acid. His-overproducing lines also displayed elevated tolerance to Ni but did not exhibit increased Ni concentrations in either xylem sap or shoot tissue, suggesting that additional factors are necessary to recapitulate the complete hyperaccumulator phenotype. These results suggest that ATP-PRT expression plays a major role in regulating the pool of free His and contributes to the exceptional Ni tolerance of hyperaccumulator Alyssum species.
Figures


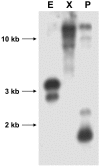
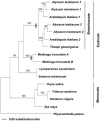

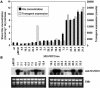
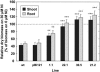


References
-
- Ames, B.N., Martin, R.G., and Garry, B.J. (1961). The first step of histidine biosynthesis. J. Biol. Chem. 236, 2019–2026. - PubMed
-
- Assunção, A.G.L., Da Costa Martins, P., De Folter, S., Vooijs, R., Schat, H., and Aarts, M.G.M. (2001). Elevated expression of metal transporter genes in three accessions of the metal hyperaccumulator Thlaspi caerulescens. Plant Cell Environ. 24, 217–226.
-
- Assunção, A.G.L., Ten Bookum, W.M., Nelissen, H.J.M., Vooijs, R., Schat, H., and Ernst, W.H.O. (2003). A cosegregation analysis of zinc (Zn) accumulation and Zn tolerance in the Zn hyperaccumulator Thlaspi caerulescens. New Phytol. 159, 383–390. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

