Mechanism of Aurora-B degradation and its dependency on intact KEN and A-boxes: identification of an aneuploidy-promoting property
- PMID: 15923616
- PMCID: PMC1140599
- DOI: 10.1128/MCB.25.12.4977-4992.2005
Mechanism of Aurora-B degradation and its dependency on intact KEN and A-boxes: identification of an aneuploidy-promoting property
Abstract
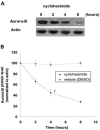
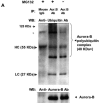
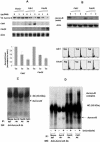
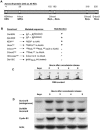
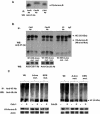
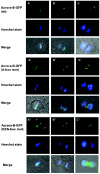
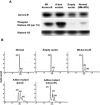
The kinase Aurora-B, a regulator of chromosome segregation and cytokinesis, is highly expressed in a variety of tumors. During the cell cycle, the level of this protein is tightly controlled, and its deregulated abundance is suspected to contribute to aneuploidy. Here, we provide evidence that Aurora-B is a short-lived protein degraded by the proteasome via the anaphase-promoting cyclosome complex (APC/c) pathway. Aurora-B interacts with the APC/c through the Cdc27 subunit, Aurora-B is ubiquitinated, and its level is increased upon treatment with inhibitors of the proteasome. Aurora-B binds in vivo to the degradation-targeting proteins Cdh1 and Cdc20, the overexpression of which accelerates Aurora-B degradation. Using deletions or point mutations of the five putative degradation signals in Aurora-B, we show that degradation of this protein does not depend on its D-boxes (RXXL), but it does require intact KEN boxes and A-boxes (QRVL) located within the first 65 amino acids. Cells transfected with wild-type or A-box-mutated or KEN box-mutated Aurora-B fused to green fluorescent protein display the protein localized to the chromosomes and then to the midzone during mitosis, but the mutated forms are detected at greater intensities. Hence, we identified the degradation pathway for Aurora-B as well as critical regions for its clearance. Intriguingly, overexpression of a stable form of Aurora-B alone induces aneuploidy and anchorage-independent growth.
Figures






Similar articles
-
Destruction box-dependent degradation of aurora B is mediated by the anaphase-promoting complex/cyclosome and Cdh1.Cancer Res. 2005 Oct 1;65(19):8730-5. doi: 10.1158/0008-5472.CAN-05-1500. Cancer Res. 2005. PMID: 16204042
-
Bub1 and aurora B cooperate to maintain BubR1-mediated inhibition of APC/CCdc20.J Cell Sci. 2005 Aug 15;118(Pt 16):3639-52. doi: 10.1242/jcs.02487. Epub 2005 Jul 26. J Cell Sci. 2005. PMID: 16046481
-
Degradation of human Aurora-A protein kinase is mediated by hCdh1.FEBS Lett. 2002 May 22;519(1-3):59-65. doi: 10.1016/s0014-5793(02)02711-4. FEBS Lett. 2002. PMID: 12023018
-
Methods to measure ubiquitin-dependent proteolysis mediated by the anaphase-promoting complex.Methods. 2006 Jan;38(1):39-51. doi: 10.1016/j.ymeth.2005.07.005. Methods. 2006. PMID: 16343932 Review.
-
The anaphase-promoting complex/cyclosome: APC/C.J Cell Sci. 2006 Jun 15;119(Pt 12):2401-4. doi: 10.1242/jcs.02937. J Cell Sci. 2006. PMID: 16763193 Review. No abstract available.
Cited by
-
Quercetin suppresses lung cancer growth by targeting Aurora B kinase.Cancer Med. 2016 Nov;5(11):3156-3165. doi: 10.1002/cam4.891. Epub 2016 Oct 5. Cancer Med. 2016. PMID: 27704720 Free PMC article.
-
The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis.Nat Rev Mol Cell Biol. 2012 Dec;13(12):789-803. doi: 10.1038/nrm3474. Nat Rev Mol Cell Biol. 2012. PMID: 23175282 Free PMC article. Review.
-
Profiling of N-acetylated protein termini provides in-depth insights into the N-terminal nature of the proteome.Mol Cell Proteomics. 2010 May;9(5):928-39. doi: 10.1074/mcp.M900463-MCP200. Epub 2010 Jan 7. Mol Cell Proteomics. 2010. PMID: 20061308 Free PMC article.
-
Functional characterization of Anaphase Promoting Complex/Cyclosome (APC/C) E3 ubiquitin ligases in tumorigenesis.Biochim Biophys Acta. 2014 Apr;1845(2):277-93. doi: 10.1016/j.bbcan.2014.02.001. Epub 2014 Feb 22. Biochim Biophys Acta. 2014. PMID: 24569229 Free PMC article. Review.
-
Co-regulation of the antagonistic RepoMan:Aurora-B pair in proliferating cells.Mol Biol Cell. 2020 Mar 15;31(6):419-438. doi: 10.1091/mbc.E19-12-0698. Epub 2020 Jan 22. Mol Biol Cell. 2020. PMID: 31967936 Free PMC article.
References
-
- Adams, R. R., M. Carmena, and W. C. Earnshaw. 2001. Chromosomal passengers and the (aurora) ABCs of mitosis. Trends Cell Biol. 11:49-54. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
