Active Src elevates the expression of beta-catenin by enhancement of cap-dependent translation
- PMID: 15923620
- PMCID: PMC1140589
- DOI: 10.1128/MCB.25.12.5031-5039.2005
Active Src elevates the expression of beta-catenin by enhancement of cap-dependent translation
Abstract
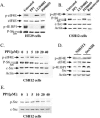
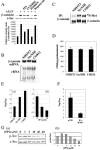
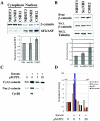
The proto-oncogene pp60(c-Src) (c-Src) is activated in many types of cancer and contributes to the transformed phenotype of the tumor, although its role is not yet fully understood. Here we report that active Src elevates the levels of beta-catenin by enhancing cap-dependent translation. Src induces phosphorylation of the eukaryotic initiation factor 4E via the Ras/Raf/ERK pathway and the phosphorylation of its inhibitor 4E-BP1 via the PI3K/mTOR pathway. Activated Src enhances the accumulation of nuclear beta-catenin and enhances its transcriptional activity, elevating target genes such as cyclin D1. This novel activation of the Wnt pathway by Src most probably contributes to the oncogenic phenotype of cancer cells.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
