Oct-3/4 maintains the proliferative embryonic stem cell state via specific binding to a variant octamer sequence in the regulatory region of the UTF1 locus
- PMID: 15923625
- PMCID: PMC1140574
- DOI: 10.1128/MCB.25.12.5084-5094.2005
Oct-3/4 maintains the proliferative embryonic stem cell state via specific binding to a variant octamer sequence in the regulatory region of the UTF1 locus
Abstract
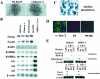
The POU transcription factor Oct-3/4 has been shown to be critical for maintaining embryonic stem (ES) cell character. However, the molecular mechanisms underlying its function remain elusive. We have previously shown that among the POU transcription factor family of proteins, Oct-3/4 alone is able to bind to the regulatory region of the UTF1 gene bearing a variant octamer sequence together with Sox-2. Here, we demonstrate using Oct-3/4-Oct-6 chimeras that there is a precise correlation between the ability of proteins to form a complex on the UTF1 enhancer with Sox-2 and the ability to maintain the stem cell state in ES cells. Different chimeric proteins show differential abilities to form a Sox-2-containing complex on the UTF1 regulatory region, with a decrease in efficiency of the complex formation accompanied by a decrease in the level of UTF1 expression and the rate of cell proliferation. Overexpression of UTF1 in these slow-growing cells was able to restore their proliferation rate to wild-type levels. Moreover, UTF1 was also observed to have an effect on teratoma formation. These results suggest a molecular pathway by which Oct-3/4 induces rapid proliferation and tumorigenic properties of ES cells through activation of the UTF1 gene.
Figures





Similar articles
-
Identification of Sox-2 regulatory region which is under the control of Oct-3/4-Sox-2 complex.Nucleic Acids Res. 2002 Jul 15;30(14):3202-13. doi: 10.1093/nar/gkf435. Nucleic Acids Res. 2002. PMID: 12136102 Free PMC article.
-
Identification of DPPA4 and other genes as putative Sox2:Oct-3/4 target genes using a combination of in silico analysis and transcription-based assays.J Cell Physiol. 2008 Sep;216(3):651-62. doi: 10.1002/jcp.21440. J Cell Physiol. 2008. PMID: 18366076
-
The gene for the embryonic stem cell coactivator UTF1 carries a regulatory element which selectively interacts with a complex composed of Oct-3/4 and Sox-2.Mol Cell Biol. 1999 Aug;19(8):5453-65. doi: 10.1128/MCB.19.8.5453. Mol Cell Biol. 1999. PMID: 10409735 Free PMC article.
-
Oct-4: gatekeeper in the beginnings of mammalian development.Stem Cells. 2001;19(4):271-8. doi: 10.1634/stemcells.19-4-271. Stem Cells. 2001. PMID: 11463946 Review.
-
POU family transcription factors in the nervous system.J Cell Physiol. 1999 May;179(2):126-33. doi: 10.1002/(SICI)1097-4652(199905)179:2<126::AID-JCP2>3.0.CO;2-M. J Cell Physiol. 1999. PMID: 10199551 Review.
Cited by
-
A unique Oct4 interface is crucial for reprogramming to pluripotency.Nat Cell Biol. 2013 Mar;15(3):295-301. doi: 10.1038/ncb2680. Epub 2013 Feb 3. Nat Cell Biol. 2013. PMID: 23376973
-
Single-cell duplex RT-LATE-PCR reveals Oct4 and Xist RNA gradients in 8-cell embryos.BMC Biotechnol. 2007 Dec 7;7:87. doi: 10.1186/1472-6750-7-87. BMC Biotechnol. 2007. PMID: 18067662 Free PMC article.
-
Biological importance of OCT transcription factors in reprogramming and development.Exp Mol Med. 2021 Jun;53(6):1018-1028. doi: 10.1038/s12276-021-00637-4. Epub 2021 Jun 11. Exp Mol Med. 2021. PMID: 34117345 Free PMC article. Review.
-
Expression profile of undifferentiated cell transcription factor 1 in normal and cancerous human epithelia.Int J Exp Pathol. 2014 Aug;95(4):251-9. doi: 10.1111/iep.12077. Epub 2014 Apr 17. Int J Exp Pathol. 2014. PMID: 24738751 Free PMC article.
-
Expression and clinical significance of undifferentiated embryonic cell transcription factor 1 in breast cancer.Transl Cancer Res. 2023 Jan 30;12(1):150-162. doi: 10.21037/tcr-22-2890. Epub 2023 Jan 13. Transl Cancer Res. 2023. PMID: 36760370 Free PMC article.
References
-
- Antonchuk, J., G. Sauvageau, and R. K. Humphries. 2002. HOXB4-induced expansion of adult hematopoietic stem cells ex vivo. Cell 109:39-45. - PubMed
-
- Ben-Shushan, E., J. R. Thompson, L. J. Gudas, and Y. Bergman. 1998. Rex-1, a gene encoding a transcription factor expressed in the early embryo, is regulated via Oct-3/4 and Oct-6 binding to an octamer site and a novel protein, Rox-1, binding to an adjacent site. Mol. Cell. Biol. 18:1866-1878. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
