Rab3D regulates a novel vesicular trafficking pathway that is required for osteoclastic bone resorption
- PMID: 15923639
- PMCID: PMC1140603
- DOI: 10.1128/MCB.25.12.5253-5269.2005
Rab3D regulates a novel vesicular trafficking pathway that is required for osteoclastic bone resorption
Abstract
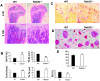
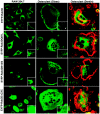
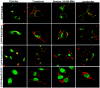
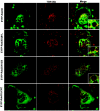
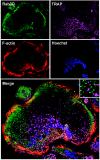
Rab3 proteins are a subfamily of GTPases, known to mediate membrane transport in eukaryotic cells and play a role in exocytosis. Our data indicate that Rab3D is the major Rab3 species expressed in osteoclasts. To investigate the role of Rab3D in osteoclast physiology we examined the skeletal architecture of Rab3D-deficient mice and found an osteosclerotic phenotype. Although basal osteoclast number in null animals is normal the total eroded surface is significantly reduced, suggesting that the resorptive defect is due to attenuated osteoclast activity. Consistent with this hypothesis, ultrastructural analysis reveals that Rab3D(-/-) osteoclasts exhibit irregular ruffled borders. Furthermore, while overexpression of wild-type, constitutively active, or prenylation-deficient Rab3D has no significant effects, overexpression of GTP-binding-deficient Rab3D impairs bone resorption in vitro. Finally, subcellular localization studies reveal that, unlike wild-type or constitutively active Rab3D, which associate with a nonendosomal/lysosomal subset of post-trans-Golgi network (TGN) vesicles, inactive Rab3D localizes to the TGN and inhibits biogenesis of Rab3D-bearing vesicles. Collectively, our data suggest that Rab3D modulates a post-TGN trafficking step that is required for osteoclastic bone resorption.
Figures







Similar articles
-
Calmodulin interacts with Rab3D and modulates osteoclastic bone resorption.Sci Rep. 2016 Nov 29;6:37963. doi: 10.1038/srep37963. Sci Rep. 2016. PMID: 27897225 Free PMC article.
-
Tctex-1, a novel interaction partner of Rab3D, is required for osteoclastic bone resorption.Mol Cell Biol. 2011 Apr;31(7):1551-64. doi: 10.1128/MCB.00834-10. Epub 2011 Jan 24. Mol Cell Biol. 2011. PMID: 21262767 Free PMC article.
-
Impaired bone resorption in cathepsin K-deficient mice is partially compensated for by enhanced osteoclastogenesis and increased expression of other proteases via an increased RANKL/OPG ratio.Bone. 2005 Jan;36(1):159-72. doi: 10.1016/j.bone.2004.09.020. Epub 2004 Nov 24. Bone. 2005. PMID: 15664014
-
Rab3D: a regulator of exocytosis in non-neuronal cells.Histol Histopathol. 2002;17(3):929-36. doi: 10.14670/HH-17.929. Histol Histopathol. 2002. PMID: 12168804 Review.
-
Rab3 proteins and cancer: Exit strategies.J Cell Biochem. 2021 Oct;122(10):1295-1301. doi: 10.1002/jcb.29948. Epub 2021 May 13. J Cell Biochem. 2021. PMID: 33982832 Review.
Cited by
-
Alcohol-Induced Liver Injury: Down-regulation and Redistribution of Rab3D Results in Atypical Protein Trafficking.Hepatol Commun. 2022 Feb;6(2):374-388. doi: 10.1002/hep4.1811. Epub 2021 Sep 7. Hepatol Commun. 2022. PMID: 34494400 Free PMC article.
-
The Roles of Small GTPases in Osteoclast Biology.Orthop Muscular Syst. 2014;3:1000161. doi: 10.4172/2161-0533.1000161. Orthop Muscular Syst. 2014. PMID: 25599004 Free PMC article.
-
Regulation of osteoclasts by membrane-derived lipid mediators.Cell Mol Life Sci. 2013 Sep;70(18):3341-53. doi: 10.1007/s00018-012-1238-4. Epub 2013 Jan 8. Cell Mol Life Sci. 2013. PMID: 23296124 Free PMC article. Review.
-
Cytoplasmic terminus of vacuolar type proton pump accessory subunit Ac45 is required for proper interaction with V(0) domain subunits and efficient osteoclastic bone resorption.J Biol Chem. 2008 May 9;283(19):13194-204. doi: 10.1074/jbc.M709712200. Epub 2008 Jan 28. J Biol Chem. 2008. PMID: 18227071 Free PMC article.
-
A Sub-Clone of RAW264.7-Cells Form Osteoclast-Like Cells Capable of Bone Resorption Faster than Parental RAW264.7 through Increased De Novo Expression and Nuclear Translocation of NFATc1.Int J Mol Sci. 2020 Jan 14;21(2):538. doi: 10.3390/ijms21020538. Int J Mol Sci. 2020. PMID: 31947698 Free PMC article.
References
-
- Abu-Amer, Y., S. L. Teitelbaum, J. C. Chappel, P. Schlesinger, and F. P. Ross. 1999. Expression and regulation of RAB3 proteins in osteoclasts and their precursors. J. Bone Miner. Res. 14:1855-1860. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
