Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages
- PMID: 15928073
- PMCID: PMC1142121
- DOI: 10.1073/pnas.0503272102
Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages
Abstract
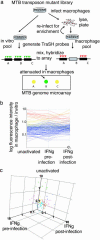
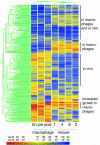
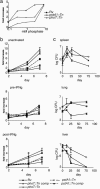
Macrophages are central to host defense against microbes, but intracellular pathogens have evolved to evade their antimicrobial functions. Mycobacterium tuberculosis (MTB) has successfully exploited macrophages as its primary niche in vivo, but the bacterial genome-wide requirements that promote its intracellular survival remain undefined. Here we comprehensively identify the MTB genes required for survival by screening for transposon mutants that fail to grow within primary macrophages. We identify mutants showing decreased growth in macrophage environments that model stages of the host immune response. By systematically analyzing several biologically relevant data sets, we have been able to identify putative pathways that could not be predicted by genome organization alone. In one example, phosphate transport, requiring physically unlinked genes, was found to be critical for MTB growth in macrophages and important for establishing persistent infection in lungs. Remarkably, the majority of MTB genes found by this analysis to be required for survival are constitutively expressed rather than regulated by macrophages, revealing the host-adapted lifestyle of an evolutionarily selected intracellular pathogen.
Figures

References
-
- Bloom, B. R. & Murray, C. J. (1992) Science 257, 1055-1064. - PubMed
-
- Flynn, J. L. & Chan, J. (2003) Curr. Opin. Immunol. 15, 450-455. - PubMed
-
- Mwandumba, H. C., Russell, D. G., Nyirenda, M. H., Anderson, J., White, S. A., Molyneux, M. E. & Squire, S. B. (2004) J. Immunol. 172, 4592-4598. - PubMed
-
- Brown, C. A., Draper, P. & Hart, P. D. (1969) Nature 221, 658-660. - PubMed
-
- Russell, D. G. (2001) Nat. Rev. Mol. Cell Biol. 2, 569-577. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

