Genomic analysis of Drosophila chromosome underreplication reveals a link between replication control and transcriptional territories
- PMID: 15928082
- PMCID: PMC1149430
- DOI: 10.1073/pnas.0502702102
Genomic analysis of Drosophila chromosome underreplication reveals a link between replication control and transcriptional territories
Abstract
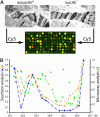
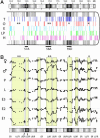
In Drosophila polytene chromosomes, most late-replicating regions remain underreplicated. A loss-of-function mutant of the suppressor of underreplication [Su(UR)] gene suppresses underreplication (UR), whereas extra copies of this gene enhance the level and number of regions showing UR. By combining DNA microarray analysis with manipulation of the number of Su(UR) gene copies, we achieved genomic-scale molecular identification of 1,036 genes that are arranged in clusters located in 52 UR chromosomal regions. These regions overlap extensively (96%) but are not completely identical with late-replicating regions of mitotically dividing Kc cells in culture. Reanalysis of published gene expression profiles revealed that genomic regions defined by replication properties include clusters of coordinately expressed genes. Genomic regions that are UR in polytene chromosomes and late replicated in Kc cell chromosomes show a particularly common association with transcriptional territories that are expressed in testis/males but not ovary/females or embryos. An attractive hypothesis for future testing is that factors involved in replication control, such as SU(UR), may interact physically with those involved in epigenetic silencing of transcription territories.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

