UNR translation can be driven by an IRES element that is negatively regulated by polypyrimidine tract binding protein
- PMID: 15928332
- PMCID: PMC1142345
- DOI: 10.1093/nar/gki611
UNR translation can be driven by an IRES element that is negatively regulated by polypyrimidine tract binding protein
Abstract
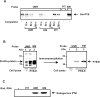
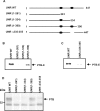
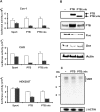
Upstream of N-ras (Unr) has been described as an internal initiation trans-acting factor (ITAF) in the cap-independent translation of some particular viral and cellular mRNAs. Two factors led us to hypothesize that the UNR 5'-untranslated region (5'-UTR) may contain an internal ribosome entry site (IRES). The first was the requirement for persisting Unr expression under conditions that correlate with cap-independent translation. The other was the observation that the primary UNR transcript contains a 447 nt long 5'-UTR including two upstream AUGs that may restrict translation initiation via cap-dependent ribosome scanning. Here we report that the UNR 5'-UTR allows IRES-dependent translation, as revealed by a dicistronic reporter assay. Various controls ruled out the contribution of leaky scanning, cryptic promoter sequences or RNA processing events to the ability of the UNR 5'-UTR to mediate internal initiation of translation. Ultraviolet cross-linking analysis and RNA affinity chromatography revealed the binding of polypyrimidine tract binding protein (PTB) to the UNR IRES, requiring a pyrimidine-rich region (nucleotides 335-355). Whereas overexpression of PTB in several cell lines inhibited UNR IRES activity and UNR protein expression, depletion of endogenous PTB using RNAi increased UNR IRES activity. Moreover, a mutant version of the UNR IRES lacking the PTB binding site was more efficient at directing IRES-mediated translation. In conclusion, our results demonstrate that translation of the ITAF Unr can itself be regulated by an IRES that is downregulated by PTB.
Figures




Similar articles
-
Polypyrimidine tract-binding protein inhibits translation of bip mRNA.J Mol Biol. 2000 Nov 24;304(2):119-33. doi: 10.1006/jmbi.2000.4179. J Mol Biol. 2000. PMID: 11080450
-
Direct evidence that polypyrimidine tract binding protein (PTB) is essential for internal initiation of translation of encephalomyocarditis virus RNA.RNA. 1995 Nov;1(9):924-38. RNA. 1995. PMID: 8548657 Free PMC article.
-
Regulation of Unr expression by 5'- and 3'-untranslated regions of its mRNA through modulation of stability and IRES mediated translation.RNA Biol. 2005 Jul-Sep;2(3):e27-35. Epub 2005 Jul 23. RNA Biol. 2005. PMID: 17114922
-
An atypical IRES within the 5' UTR of a dicistrovirus genome.Virus Res. 2009 Feb;139(2):157-65. doi: 10.1016/j.virusres.2008.07.017. Epub 2008 Sep 11. Virus Res. 2009. PMID: 18755228 Review.
-
Mechanism of translation initiation on hepatitis C virus RNA.Princess Takamatsu Symp. 1995;25:111-9. Princess Takamatsu Symp. 1995. PMID: 8875615 Review.
Cited by
-
CSDE1 attenuates microRNA-mediated silencing of PMEPA1 in melanoma.Oncogene. 2021 May;40(18):3231-3244. doi: 10.1038/s41388-021-01767-9. Epub 2021 Apr 8. Oncogene. 2021. PMID: 33833398
-
RNA Binding Proteins and Regulation of mRNA Translation in Erythropoiesis.Front Physiol. 2018 Jul 24;9:910. doi: 10.3389/fphys.2018.00910. eCollection 2018. Front Physiol. 2018. PMID: 30087616 Free PMC article. Review.
-
Searching for IRES.RNA. 2006 Oct;12(10):1755-85. doi: 10.1261/rna.157806. Epub 2006 Sep 6. RNA. 2006. PMID: 16957278 Free PMC article. Review.
-
The hnRNA-binding proteins hnRNP L and PTB are required for efficient translation of the Cat-1 arginine/lysine transporter mRNA during amino acid starvation.Mol Cell Biol. 2009 May;29(10):2899-912. doi: 10.1128/MCB.01774-08. Epub 2009 Mar 9. Mol Cell Biol. 2009. PMID: 19273590 Free PMC article.
-
A deep learning framework for modeling structural features of RNA-binding protein targets.Nucleic Acids Res. 2016 Feb 29;44(4):e32. doi: 10.1093/nar/gkv1025. Epub 2015 Oct 13. Nucleic Acids Res. 2016. PMID: 26467480 Free PMC article.
References
-
- Stoneley M., Willis A.E. Cellular internal ribosome entry segments: structures, trans-acting factors and regulation of gene expression. Oncogene. 2004;23:3200–3207. - PubMed
-
- Fernandez J., Yaman I., Mishra R., Merrick W.C., Snider M.D., Lamers W.H., Hatzoglou M. Internal ribosome entry site-mediated translation of a mammalian mRNA is regulated by amino acid availability. J. Biol. Chem. 2001;276:12285–12291. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

