Weighted sequence motifs as an improved seeding step in microRNA target prediction algorithms
- PMID: 15928346
- PMCID: PMC1370784
- DOI: 10.1261/rna.7290705
Weighted sequence motifs as an improved seeding step in microRNA target prediction algorithms
Abstract
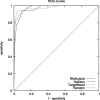
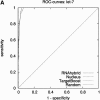
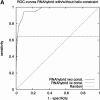
We present a new microRNA target prediction algorithm called TargetBoost, and show that the algorithm is stable and identifies more true targets than do existing algorithms. TargetBoost uses machine learning on a set of validated microRNA targets in lower organisms to create weighted sequence motifs that capture the binding characteristics between microRNAs and their targets. Existing algorithms require candidates to have (1) near-perfect complementarity between microRNAs' 5' end and their targets; (2) relatively high thermodynamic duplex stability; (3) multiple target sites in the target's 3' UTR; and (4) evolutionary conservation of the target between species. Most algorithms use one of the two first requirements in a seeding step, and use the three others as filters to improve the method's specificity. The initial seeding step determines an algorithm's sensitivity and also influences its specificity. As all algorithms may add filters to increase the specificity, we propose that methods should be compared before such filtering. We show that TargetBoost's weighted sequence motif approach is favorable to using both the duplex stability and the sequence complementarity steps. (TargetBoost is available as a Web tool from http://www.interagon.com/demo/.).
Figures








Similar articles
-
Fast and effective prediction of microRNA/target duplexes.RNA. 2004 Oct;10(10):1507-17. doi: 10.1261/rna.5248604. RNA. 2004. PMID: 15383676 Free PMC article.
-
Analysis of microRNA-target interactions by a target structure based hybridization model.Pac Symp Biocomput. 2008:64-74. Pac Symp Biocomput. 2008. PMID: 18232104
-
The role of site accessibility in microRNA target recognition.Nat Genet. 2007 Oct;39(10):1278-84. doi: 10.1038/ng2135. Epub 2007 Sep 23. Nat Genet. 2007. PMID: 17893677
-
Computational Prediction of MicroRNA Target Genes, Target Prediction Databases, and Web Resources.Methods Mol Biol. 2017;1617:109-122. doi: 10.1007/978-1-4939-7046-9_8. Methods Mol Biol. 2017. PMID: 28540680 Review.
-
Prediction and validation of microRNAs and their targets.FEBS Lett. 2005 Oct 31;579(26):5904-10. doi: 10.1016/j.febslet.2005.09.040. Epub 2005 Sep 30. FEBS Lett. 2005. PMID: 16214134 Review.
Cited by
-
Comprehensive analysis of microRNA genomic loci identifies pervasive repetitive-element origins.Mob Genet Elements. 2011 May;1(1):8-17. doi: 10.4161/mge.1.1.15766. Mob Genet Elements. 2011. PMID: 22016841 Free PMC article.
-
Hsa-miR-520d converts fibroblasts into CD105+ populations.Drugs R D. 2014 Dec;14(4):253-64. doi: 10.1007/s40268-014-0064-6. Drugs R D. 2014. PMID: 25303886 Free PMC article.
-
Toward a complete in silico, multi-layered embryonic stem cell regulatory network.Wiley Interdiscip Rev Syst Biol Med. 2010 Nov-Dec;2(6):708-33. doi: 10.1002/wsbm.93. Wiley Interdiscip Rev Syst Biol Med. 2010. PMID: 20890967 Free PMC article. Review.
-
A Bayesian approach for identifying miRNA targets by combining sequence prediction and gene expression profiling.BMC Genomics. 2010 Dec 1;11 Suppl 3(Suppl 3):S12. doi: 10.1186/1471-2164-11-S3-S12. BMC Genomics. 2010. PMID: 21143779 Free PMC article.
-
Current tools for the identification of miRNA genes and their targets.Nucleic Acids Res. 2009 May;37(8):2419-33. doi: 10.1093/nar/gkp145. Epub 2009 Mar 18. Nucleic Acids Res. 2009. PMID: 19295136 Free PMC article. Review.
References
-
- Bartel, D.P. 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297. - PubMed
-
- Breiman, L., Friedman, J.H., Olshen, R.A., and Stone, C.J. 1984. Classification and regression trees. Wadsworth, Belmont, CA.
-
- Brennecke, J., Hipfner, D.R., Stark, A., Russell, R.B., and Cohen, S.M. 2003. bantam Encodes a developmentally regulated miRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113: 25–36. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
