Deletion of cagA gene of Helicobacter pylori by PCR products
- PMID: 15929177
- PMCID: PMC4316058
- DOI: 10.3748/wjg.v11.i21.3255
Deletion of cagA gene of Helicobacter pylori by PCR products
Abstract
Aim: Cytotoxin-associated protein (antigen) A (CagA) plays an important role in Helicobacter pylori (H pylori) pathogenesis. Our aim was to obtain cagA- mutant strains by a new mutation method so as to better understand the mechanism of CagA in epithelial cells.
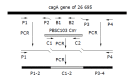
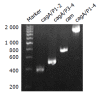
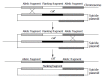
Methods: In contrast with the traditional method using suicide plasmid, we constructed cagA- mutant strains directly with PCR products. The constructed mutant clones grew on selective media and allelic exchange was confirmed by Southern blot. Furthermore, two different transformation methods, electroporation, and natural transformation, were compared with regard to the efficiency of recombination.
Results: The mutation by PCR products could be completed within 3-5 d, and the recombination rate by electroporation and natural transformation was 4.02 x 10(-8) and 1.03 x 10(-9) respectively. Mutation rate by electroporation (4.02 x 10(-8)) was far higher than by natural transformation (1.03 x 10(-9)) (P = 0.000<0.005).
Conclusion: cagA- mutant strains have been constructed, which is important for further study on the function of CagA in epithelial cells. A mutation method by directly using PCR products has been proved successful with a much higher mutation rate, and is easier, especially when in combination with electroporation. This method could be widely used in gene deletion of H pylori.
Figures

Similar articles
-
[Construction of hp0523 gene mutant in Helicobacter pylori Cag-PAI and the influence on the ability of CagA protein translocation].Wei Sheng Wu Xue Bao. 2009 Apr;49(4):460-4. Wei Sheng Wu Xue Bao. 2009. PMID: 19621632 Chinese.
-
A mutagenesis method for the addition and deletion of highly repetitive DNA regions: the paradigm of EPIYA motifs in the cagA gene of Helicobacter pylori.Helicobacter. 2013 Jun;18(3):229-41. doi: 10.1111/hel.12029. Epub 2012 Nov 27. Helicobacter. 2013. PMID: 23190444
-
[Detection of cagA prevalence in clinical isolates of Helicobacter pylori].Mikrobiyol Bul. 2010 Jul;44(3):461-5. Mikrobiyol Bul. 2010. PMID: 21063996 Turkish.
-
Host cell contact induces fur-dependent expression of virulence factors CagA and VacA in Helicobacter pylori.Helicobacter. 2014 Feb;19(1):17-25. doi: 10.1111/hel.12087. Epub 2013 Sep 11. Helicobacter. 2014. PMID: 24020886
-
[Advances in CagA protein and CagAmediated pathogenesis of Helicobacter pylori-A review].Wei Sheng Wu Xue Bao. 2016 Dec 4;56(12):1821-30. Wei Sheng Wu Xue Bao. 2016. PMID: 29741846 Review. Chinese.
Cited by
-
A novel insight into the oxidoreductase activity of Helicobacter pylori HP0231 protein.PLoS One. 2012;7(10):e46563. doi: 10.1371/journal.pone.0046563. Epub 2012 Oct 3. PLoS One. 2012. PMID: 23056345 Free PMC article.
-
Adherence and invasion of mouse-adapted H pylori in different epithelial cell lines.World J Gastroenterol. 2007 Feb 14;13(6):845-50. doi: 10.3748/wjg.v13.i6.845. World J Gastroenterol. 2007. PMID: 17352012 Free PMC article.
-
N4-cytosine DNA methylation regulates transcription and pathogenesis in Helicobacter pylori.Nucleic Acids Res. 2018 Apr 20;46(7):3429-3445. doi: 10.1093/nar/gky126. Nucleic Acids Res. 2018. PMID: 29481677 Free PMC article.
-
Helicobacter pylori HP0377, a member of the Dsb family, is an untypical multifunctional CcmG that cooperates with dimeric thioldisulfide oxidase HP0231.BMC Microbiol. 2015 Jul 4;15:135. doi: 10.1186/s12866-015-0471-z. BMC Microbiol. 2015. PMID: 26141380 Free PMC article.
References
-
- Covacci A, Telford JL, Del Giudice G, Parsonnet J, Rappuoli R. Helicobacter pylori virulence and genetic geography. Science. 1999;284:1328–1333. - PubMed
-
- Marchetti M, Aricò B, Burroni D, Figura N, Rappuoli R, Ghiara P. Development of a mouse model of Helicobacter pylori infection that mimics human disease. Science. 1995;267:1655–1658. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

