Bacterial activity in cystic fibrosis lung infections
- PMID: 15929792
- PMCID: PMC1177990
- DOI: 10.1186/1465-9921-6-49
Bacterial activity in cystic fibrosis lung infections
Abstract
Background: Chronic lung infections are the primary cause of morbidity and mortality in Cystic Fibrosis (CF) patients. Recent molecular biological based studies have identified a surprisingly wide range of hitherto unreported bacterial species in the lungs of CF patients. The aim of this study was to determine whether the species present were active and, as such, worthy of further investigation as potential pathogens.
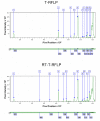
Methods: Terminal Restriction Fragment Length Polymorphism (T-RFLP) profiles were generated from PCR products amplified from 16S rDNA and Reverse Transcription Terminal Restriction Fragment Length Polymorphism (RT-T-RFLP) profiles, a marker of metabolic activity, were generated from PCR products amplified from 16S rRNA, both extracted from the same CF sputum sample. To test the level of activity of these bacteria, T-RFLP profiles were compared to RT-T-RFLP profiles.
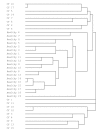
Results: Samples from 17 individuals were studied. Parallel analyses identified a total of 706 individual T-RF and RT-T-RF bands in this sample set. 323 bands were detected by T-RFLP and 383 bands were detected by RT-T-RFLP (statistically significant; P < or = 0.001). For the group as a whole, 145 bands were detected in a T-RFLP profile alone, suggesting metabolically inactive bacteria. 205 bands were detected in an RT-T-RFLP profile alone and 178 bands were detected in both, suggesting a significant degree of metabolic activity. Although Pseudomonas aeruginosa was present and active in many patients, a low occurrence of other species traditionally considered to be key CF pathogens was detected. T-RFLP profiles obtained for induced sputum samples provided by healthy individuals without CF formed a separate cluster indicating a low level of similarity to those from CF patients.
Conclusion: These results indicate that a high proportion of the bacterial species detected in the sputum from all of the CF patients in the study are active. The widespread activity of bacterial species in these samples emphasizes the potential importance of these previously unrecognized species within the CF lung.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

