Dichotomous splicing signals in exon flanks
- PMID: 15930489
- PMCID: PMC1142467
- DOI: 10.1101/gr.3217705
Dichotomous splicing signals in exon flanks
Abstract
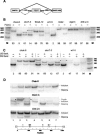
Intronic elements flanking the splice-site consensus sequences are thought to play a role in pre-mRNA splicing. However, the generality of this role, the catalog of effective sequences, and the mechanisms involved are still lacking. Using molecular genetic tests, we first showed that the approximately 50-nt intronic flanking sequences of exons beyond the splice-site consensus are generally important for splicing. We then went on to characterize exon flank sequences on a genomic scale. The G+C content of flanks displayed a bimodal distribution reflecting an exaggeration of this base composition in flanks relative to the gene as a whole. We divided all exons into two classes according to their flank G+C content and used computational and statistical methods to define pentamers of high relative abundance and phylogenetic conservation in exon flanks. Upstream pentamers were often common to the two classes, whereas downstream pentamers were totally different. Upstream and downstream pentamers were often identical around low G+C exons, and in contrast, were often complementary around high G+C exons. In agreement with this complementarity, predicted base pairing was more frequent between the flanks of high G+C exons. Pseudo exons did not exhibit this behavior, but rather tended to form base pairs between flanks and exon bodies. We conclude that most exons require signals in their immediate flanks for efficient splicing. G+C content is a sequence feature correlated with many genetic and genomic attributes. We speculate that there may be different mechanisms for splice site recognition depending on G+C content.
Figures





Similar articles
-
Intronic motif pairs cooperate across exons to promote pre-mRNA splicing.Genome Biol. 2010;11(8):R84. doi: 10.1186/gb-2010-11-8-r84. Epub 2010 Aug 12. Genome Biol. 2010. PMID: 20704715 Free PMC article.
-
Sequence information for the splicing of human pre-mRNA identified by support vector machine classification.Genome Res. 2003 Dec;13(12):2637-50. doi: 10.1101/gr.1679003. Genome Res. 2003. PMID: 14656968 Free PMC article.
-
Compensatory relationship between splice sites and exonic splicing signals depending on the length of vertebrate introns.BMC Genomics. 2006 Dec 8;7:311. doi: 10.1186/1471-2164-7-311. BMC Genomics. 2006. PMID: 17156453 Free PMC article.
-
Exonization of transposed elements: A challenge and opportunity for evolution.Biochimie. 2011 Nov;93(11):1928-34. doi: 10.1016/j.biochi.2011.07.014. Epub 2011 Jul 26. Biochimie. 2011. PMID: 21787833 Review.
-
Searching for splicing motifs.Adv Exp Med Biol. 2007;623:85-106. doi: 10.1007/978-0-387-77374-2_6. Adv Exp Med Biol. 2007. PMID: 18380342 Review.
Cited by
-
Splicing of a critical exon of human Survival Motor Neuron is regulated by a unique silencer element located in the last intron.Mol Cell Biol. 2006 Feb;26(4):1333-46. doi: 10.1128/MCB.26.4.1333-1346.2006. Mol Cell Biol. 2006. PMID: 16449646 Free PMC article.
-
Intronic motif pairs cooperate across exons to promote pre-mRNA splicing.Genome Biol. 2010;11(8):R84. doi: 10.1186/gb-2010-11-8-r84. Epub 2010 Aug 12. Genome Biol. 2010. PMID: 20704715 Free PMC article.
-
A systematic analysis of intronic sequences downstream of 5' splice sites reveals a widespread role for U-rich motifs and TIA1/TIAL1 proteins in alternative splicing regulation.Genome Res. 2008 Aug;18(8):1247-58. doi: 10.1101/gr.073155.107. Epub 2008 May 2. Genome Res. 2008. PMID: 18456862 Free PMC article.
-
GHR gene transcript heterogeneity may explain phenotypic variability in GHR pseudoexon (6Ψ) patients.Endocr Connect. 2020 Mar;9(3):211-222. doi: 10.1530/EC-20-0026. Endocr Connect. 2020. PMID: 32061156 Free PMC article.
-
A functional conserved intronic G run in HIV-1 intron 3 is critical to counteract APOBEC3G-mediated host restriction.Retrovirology. 2014 Aug 29;11:72. doi: 10.1186/s12977-014-0072-1. Retrovirology. 2014. PMID: 25169827 Free PMC article.
References
-
- Abdul-Manan, N., O'Malley, S.M., and Williams, K.R. 1996. Origins of binding specificity of the A1 heterogeneous nuclear ribonucleoprotein. Biochemistry 35: 3545-3554. - PubMed
-
- Adams, M.D., Rudner, D.Z., and Rio, D.C. 1996. Biochemistry and regulation of pre-mRNA splicing. Curr. Opin. Cell. Biol. 8: 331-339. - PubMed
-
- Berget, S.M. 1995. Exon recognition in vertebrate splicing. J. Biol. Chem. 270: 2411-2414. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
