The genetic epidemiology of neurodegenerative disease
- PMID: 15931380
- PMCID: PMC1137006
- DOI: 10.1172/JCI24761
The genetic epidemiology of neurodegenerative disease
Abstract
Gene defects play a major role in the pathogenesis of degenerative disorders of the nervous system. In fact, it has been the very knowledge gained from genetic studies that has allowed the elucidation of the molecular mechanisms underlying the etiology and pathogenesis of many neurodegenerative disorders. In this review, we discuss the current status of genetic epidemiology of the most common neurodegenerative diseases: Alzheimer disease, Parkinson disease, Lewy body dementia, frontotemporal dementia, amyotrophic lateral sclerosis, Huntington disease, and prion diseases, with a particular focus on similarities and differences among these syndromes.
Figures
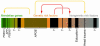
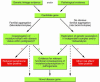

References
-
- Lander ES. The new genomics: global views of biology. Science. 1996;274:536–539. - PubMed
-
- Risch NJ. Searching for genetic determinants in the new millennium. Nature. 2000;405:847–856. - PubMed
-
- Hardy J, Myers A, Wavrant-De Vriese F. Problems and solutions in the genetic analysis of late-onset Alzheimer’s disease. Neurodegenerative Diseases. 2004;1:213–217. - PubMed
-
- Munafo MR, Clark TG, Flint J. Assessing publication bias in genetic association studies: evidence from a recent meta-analysis. Psychiatry Res. 2004;129:39–44. - PubMed
-
- Colhoun HM, McKeigue PM, Davey Smith G. Problems of reporting genetic associations with complex outcomes. Lancet. 2003;361:865–872. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

