Characterization of a nucleus-encoded chitinase from the yeast Kluyveromyces lactis
- PMID: 15932978
- PMCID: PMC1151841
- DOI: 10.1128/AEM.71.6.2862-2869.2005
Characterization of a nucleus-encoded chitinase from the yeast Kluyveromyces lactis
Abstract
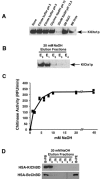
Endogenous proteins secreted from Kluyveromyces lactis were screened for their ability to bind to or to hydrolyze chitin. This analysis resulted in identification of a nucleus-encoded extracellular chitinase (KlCts1p) with a chitinolytic activity distinct from that of the plasmid-encoded killer toxin alpha-subunit. Sequence analysis of cloned KlCTS1 indicated that it encodes a 551-amino-acid chitinase having a secretion signal peptide, an amino-terminal family 18 chitinase catalytic domain, a serine-threonine-rich domain, and a carboxy-terminal type 2 chitin-binding domain. The association of purified KlCts1p with chitin is stable in the presence of high salt concentrations and pH 3 to 10 buffers; however, complete dissociation and release of fully active KlCts1p occur in 20 mM NaOH. Similarly, secreted human serum albumin harboring a carboxy-terminal fusion with the chitin-binding domain derived from KlCts1p also dissociates from chitin in 20 mM NaOH, demonstrating the domain's potential utility as an affinity tag for reversible chitin immobilization or purification of alkaliphilic or alkali-tolerant recombinant fusion proteins. Finally, haploid K. lactis cells harboring a cts1 null mutation are viable but exhibit a cell separation defect, suggesting that KlCts1p is required for normal cytokinesis, probably by facilitating the degradation of septum-localized chitin.
Figures





References
-
- Arakane, Y., Q. Zhu, M. Matsumiya, S. Muthukrishnan, and K. J. Kramer. 2003. Properties of catalytic, linker and chitin-binding domains of insect chitinase. Insect Biochem. Mol. Biol. 33:631-648. - PubMed
-
- Asensio, J. L., F. J.Canada, H. C. Siebert, J. Laynez, A. Poveda, P. M. Nieto, U. M. Soedjanaamadja, H. J. Gabius, and J. Jimenez-Barbero. 2000. Structural basis for chitin recognition by defense proteins: GlcNAc residues are bound in a multivalent fashion by extended binding sites in hevein domains. Chem. Biol. 7:529-543. - PubMed
-
- Bernard, M. P., D. Cao, R. V. Myers, and W. R. Moyle. 2004. Tight attachment of chitin-binding-domain-tagged proteins to surfaces coated with acetylated chitosan. Anal. Biochem. 327:278-283. - PubMed
-
- Bokma, E., H. J. Rozeboom, M. Sibbald, B. W. Dijkstra, and J. J. Beintema. 2002. Expression and characterization of active site mutants of hevamine, a chitinase from the rubber tree Hevea brasiliensis. Eur. J. Biochem. 269:893-901. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

