Accelerated sulfur cycle in coastal marine sediment beneath areas of intensive shellfish aquaculture
- PMID: 15932986
- PMCID: PMC1151846
- DOI: 10.1128/AEM.71.6.2925-2933.2005
Accelerated sulfur cycle in coastal marine sediment beneath areas of intensive shellfish aquaculture
Abstract
Prokaryotes in marine sediments taken from two neighboring semi-enclosed bays (the Yamada and Kamaishi bays) at the Sanriku coast in Japan were investigated by the culture-independent molecular phylogenetic approach coupled with chemical and activity analyses. These two bays were chosen in terms of their similar hydrogeological and chemical characteristics but different usage modes; the Yamada bay has been used for intensive shellfish aquaculture, while the Kamaishi bay has a commercial port and is not used for aquaculture. Substantial differences were found in the phylogenetic composition of 16S rRNA gene clone libraries constructed for the Yamada and Kamaishi sediments. In the Yamada library, phylotypes affiliated with delta-Proteobacteria were the most abundant, and those affiliated with gamma-Proteobacteria were the second-most abundant. In contrast, the Kamaishi library was occupied by phylotypes affiliated with Planctomycetes, gamma-Proteobacteria, delta-Proteobacteria, and Crenarchaeota. In the gamma-Proteobacteria, many Yamada phylotypes were related to free-living and symbiotic sulfur oxidizers, whereas the Kamaishi phylotype was related to the genus Pseudomonas. These results allowed us to hypothesize that sulfate-reducing and sulfur-oxidizing bacteria have become abundant in the Yamada sediment. This hypothesis was supported by quantitative competitive PCR (qcPCR) with group-specific primers. The qcPCR also suggested that organisms closely related to Desulfotalea in the Desulfobulbaceae were the major sulfate-reducing bacteria in these sediments. In addition, potential sulfate reduction and sulfur oxidation rates in the sediment samples were determined, indicating that the sulfur cycle has become active in the Yamada sediment beneath the areas of intensive shellfish aquaculture.
Figures



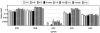
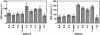
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

