An ergot alkaloid biosynthesis gene and clustered hypothetical genes from Aspergillus fumigatus
- PMID: 15933009
- PMCID: PMC1151871
- DOI: 10.1128/AEM.71.6.3112-3118.2005
An ergot alkaloid biosynthesis gene and clustered hypothetical genes from Aspergillus fumigatus
Abstract
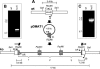
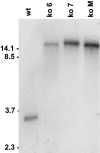
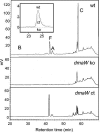
The ergot alkaloids are a family of indole-derived mycotoxins with a variety of significant biological activities. Aspergillus fumigatus, a common airborne fungus and opportunistic human pathogen, and several fungi in the relatively distant taxon Clavicipitaceae (clavicipitaceous fungi) produce different sets of ergot alkaloids. The ergot alkaloids of these divergent fungi share a four-member ergoline ring but differ in the number, type, and position of the side chains. Several genes required for ergot alkaloid production are known in the clavicipitaceous fungi, and these genes are clustered in the genome of the ergot fungus Claviceps purpurea. We investigated whether the ergot alkaloids of A. fumigatus have a common biosynthetic and genetic origin with those of the clavicipitaceous fungi. A homolog of dmaW, the gene controlling the determinant step in the ergot alkaloid pathway of clavicipitaceous fungi, was identified in the A. fumigatus genome. Knockout of dmaW eliminated all known ergot alkaloids from A. fumigatus, and complementation of the mutation restored ergot alkaloid production. Clustered with dmaW in the A. fumigatus genome are sequences corresponding to five genes previously proposed to encode steps in the ergot alkaloid pathway of C. purpurea, as well as additional sequences whose deduced protein products are consistent with their involvement in the ergot alkaloid pathway. The corresponding genes have similarities in their nucleotide sequences, but the orientations and positions within the cluster of several of these genes differ. The data indicate that the ergot alkaloid biosynthetic capabilities in A. fumigatus and the clavicipitaceous fungi had a common origin.
Figures

Similar articles
-
Ergot cluster-encoded catalase is required for synthesis of chanoclavine-I in Aspergillus fumigatus.Curr Genet. 2011 Jun;57(3):201-11. doi: 10.1007/s00294-011-0336-4. Epub 2011 Mar 17. Curr Genet. 2011. PMID: 21409592
-
An old yellow enzyme gene controls the branch point between Aspergillus fumigatus and Claviceps purpurea ergot alkaloid pathways.Appl Environ Microbiol. 2010 Jun;76(12):3898-903. doi: 10.1128/AEM.02914-09. Epub 2010 Apr 30. Appl Environ Microbiol. 2010. PMID: 20435769 Free PMC article.
-
Post-genome research on the biosynthesis of ergot alkaloids.Planta Med. 2006 Oct;72(12):1117-20. doi: 10.1055/s-2006-947195. Epub 2006 Aug 10. Planta Med. 2006. PMID: 16902860 Review.
-
Evidence for an ergot alkaloid gene cluster in Claviceps purpurea.Mol Gen Genet. 1999 Feb;261(1):133-41. doi: 10.1007/s004380050950. Mol Gen Genet. 1999. PMID: 10071219
-
Ergot alkaloids--biology and molecular biology.Alkaloids Chem Biol. 2006;63:45-86. doi: 10.1016/s1099-4831(06)63002-2. Alkaloids Chem Biol. 2006. PMID: 17133714 Review.
Cited by
-
Copper starvation induces antimicrobial isocyanide integrated into two distinct biosynthetic pathways in fungi.Nat Commun. 2022 Aug 16;13(1):4828. doi: 10.1038/s41467-022-32394-x. Nat Commun. 2022. PMID: 35973982 Free PMC article.
-
Nonribosomal peptide synthetase genes pesL and pes1 are essential for Fumigaclavine C production in Aspergillus fumigatus.Appl Environ Microbiol. 2012 May;78(9):3166-76. doi: 10.1128/AEM.07249-11. Epub 2012 Feb 17. Appl Environ Microbiol. 2012. PMID: 22344643 Free PMC article.
-
Non-Native Site-Selective Enzyme Catalysis.Chem Rev. 2023 Aug 23;123(16):10381-10431. doi: 10.1021/acs.chemrev.3c00215. Epub 2023 Jul 31. Chem Rev. 2023. PMID: 37524057 Free PMC article. Review.
-
Ergot cluster-encoded catalase is required for synthesis of chanoclavine-I in Aspergillus fumigatus.Curr Genet. 2011 Jun;57(3):201-11. doi: 10.1007/s00294-011-0336-4. Epub 2011 Mar 17. Curr Genet. 2011. PMID: 21409592
-
A Timeline of Biosynthetic Gene Cluster Discovery in Aspergillus fumigatus: From Characterization to Future Perspectives.J Fungi (Basel). 2024 Apr 2;10(4):266. doi: 10.3390/jof10040266. J Fungi (Basel). 2024. PMID: 38667937 Free PMC article.
References
-
- Apel, P. A., D. G. Panaccione, F. R. Holden, and J. D. Walton. 1993. Cloning and targeted gene disruption of XYL1, a β1,4-xylanase gene from the maize pathogen Cochliobolus carbonum. Mol. Plant-Microbe Interact. 6:467-473. - PubMed
-
- Clay, K., and C. Schardl. 2002. Evolutionary origins and ecological consequences of endophyte symbiosis with grasses. Am. Nat. 160:S199-S127. - PubMed
-
- Cole, R. J., J. W. Kirksey, J. W. Dorner, D. M. Wilson, J. C. Johnson, Jr., A. N. Johnson, D. M. Bedell, J. P. Springer, K. K. Chexal, J. C. Clardy, and R. H. Cox. 1977. Mycotoxins produced by Aspergillus fumigatus species isolated from molded silage. J. Agric. Food Chem. 25:826-830. - PubMed
-
- Correia, T., N. Grammel, I. Ortel, U. Keller, and P. Tudzynski. 2003. Molecular cloning and analysis of the ergopeptine assembly system in the ergot fungus Claviceps purpurea. Chem. Biol. 10:1281-1292. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

