Host distributions of uncultivated fecal Bacteroidales bacteria reveal genetic markers for fecal source identification
- PMID: 15933020
- PMCID: PMC1151806
- DOI: 10.1128/AEM.71.6.3184-3191.2005
Host distributions of uncultivated fecal Bacteroidales bacteria reveal genetic markers for fecal source identification
Abstract
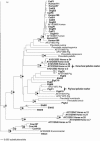
The purpose of this study was to examine host distribution patterns among fecal bacteria in the order Bacteroidales, with the goal of using endemic sequences as markers for fecal source identification in aquatic environments. We analyzed Bacteroidales 16S rRNA gene sequences from the feces of eight hosts: human, bovine, pig, horse, dog, cat, gull, and elk. Recovered sequences did not match database sequences, indicating high levels of uncultivated diversity. The analysis revealed both endemic and cosmopolitan distributions among the eight hosts. Ruminant, pig, and horse sequences tended to form host- or host group-specific clusters in a phylogenetic tree, while human, dog, cat, and gull sequences clustered together almost exclusively. Many of the human, dog, cat, and gull sequences fell within a large branch containing cultivated species from the genus Bacteroides. Most of the cultivated Bacteroides species had very close matches with multiple hosts and thus may not be useful targets for fecal source identification. A large branch containing cultivated members of the genus Prevotella included cloned sequences that were not closely related to cultivated Prevotella species. Most ruminant sequences formed clusters separate from the branches containing Bacteroides and Prevotella species. Host-specific sequences were identified for pigs and horses and were used to design PCR primers to identify pig and horse sources of fecal pollution in water. The primers successfully amplified fecal DNAs from their target hosts and did not amplify fecal DNAs from other species. Fecal bacteria endemic to the host species may result from evolution in different types of digestive systems.
Figures




References
-
- Acinas, S. G., V. Klepac-Ceraf, D. E. Hunt, C. Pharino, I. Ceraj, D. L. Distel, and M. F. Polz. 2004. Fine-scale phylogenetic architecture of a complex bacterial community. Nature 430:551-554. - PubMed
-
- Allsop, K., and J. D. Stickler. 1985. An assessment of Bacteroides fragilis group organisms as indicators of human faecal pollution. J. Appl. Bacteriol. 58:95-99. - PubMed
-
- Boone, D. R., and R. W. Castenholz (ed.). 2001. Bergey's manual of systematic bacteriology, 2nd ed., vol. 1. Springer-Verlag, New York, N.Y.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

