Rational design of a Corynebacterium glutamicum pantothenate production strain and its characterization by metabolic flux analysis and genome-wide transcriptional profiling
- PMID: 15933028
- PMCID: PMC1151861
- DOI: 10.1128/AEM.71.6.3255-3268.2005
Rational design of a Corynebacterium glutamicum pantothenate production strain and its characterization by metabolic flux analysis and genome-wide transcriptional profiling
Abstract
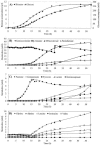
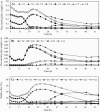
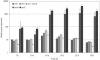
A "second-generation" production strain was derived from a Corynebacterium glutamicum pantothenate producer by rational design to assess its potential to synthesize and accumulate the vitamin pantothenate by batch cultivation. The new pantothenate production strain carries a deletion of the ilvA gene to abolish isoleucine synthesis, the promoter down-mutation P-ilvEM3 to attenuate ilvE gene expression and thereby increase ketoisovalerate availability, and two compatible plasmids to overexpress the ilvBNCD genes and duplicated copies of the panBC operon. Production assays in shake flasks revealed that the P-ilvEM3 mutation and the duplication of the panBC operon had cumulative effects on pantothenate production. During pH-regulated batch cultivation, accumulation of 8 mM pantothenate was achieved, which is the highest value reported for C. glutamicum. Metabolic flux analysis during the fermentation demonstrated that the P-ilvEM3 mutation successfully reoriented the carbon flux towards pantothenate biosynthesis. Despite this repartition of the carbon flux, ketoisovalerate not converted to pantothenate was excreted by the cell and dissipated as by-products (ketoisocaproate, DL-2,3,-dihydroxy-isovalerate, ketopantoate, pantoate), which are indicative of saturation of the pantothenate biosynthetic pathway. Genome-wide expression analysis of the production strain during batch cultivation was performed by whole-genome DNA microarray hybridization and agglomerative hierarchical clustering, which detected the enhanced expression of genes involved in leucine biosynthesis, in serine and glycine formation, in regeneration of methylenetetrahydrofolate, in de novo synthesis of nicotinic acid mononucleotide, and in a complete pathway of acyl coenzyme A conversion. Our strategy not only successfully improved pantothenate production by genetically modified C. glutamicum strains but also revealed new constraints in attaining high productivity.
Figures



References
-
- Bonamy, C., A. Guyonvarch, O. Reyes, F. David, and G. Leblon. 1990. Interspecies electro-transformation in corynebacteria. FEMS Microbiol. Lett. 66:263-269. - PubMed
-
- Brosius, J. 1988. Expression vectors employing lambda-, trp-, lac- and lpp-derived promoters, p. 205-225. In R. L. Rodriguez and D. T. Denhardt (ed.), Vectors: a survey of molecular cloning vectors and their uses. Butterworth, Boston, MA. - PubMed
-
- Chassagnole, C., F. Létisse, A. Diano, and N. D. Lindley. 2002. Carbon flux analysis in a pantothenate overproducing Corynebacterium glutamicum strain. Mol. Biol. Rep. 29:129-134. - PubMed
-
- Chassagnole, C., A. Diano, F. Létisse, and N. D. Lindley. 2003. Metabolic network analysis during fed-batch cultivation of Corynebacterium glutamicum for pantothenic acid production: first quantitative data and analysis of by-product formation. J. Biotechnol. 104:261-272. - PubMed
-
- Cordes, C., B. Möckel, L. Eggeling, and H. Sahm. 1992. Cloning, organization and functional analysis of ilvA, ilvB and ilvC genes from Corynebacterium glutamicum. Gene 112:113-116. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

