Structural and functional analysis of SET8, a histone H4 Lys-20 methyltransferase
- PMID: 15933070
- PMCID: PMC1151662
- DOI: 10.1101/gad.1318405
Structural and functional analysis of SET8, a histone H4 Lys-20 methyltransferase
Abstract
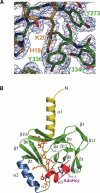
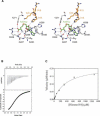
SET8 (also known as PR-SET7) is a histone H4-Lys-20-specific methyltransferase that is implicated in cell-cycle-dependent transcriptional silencing and mitotic regulation in metazoans. Herein we report the crystal structure of human SET8 (hSET8) bound to a histone H4 peptide bearing Lys-20 and the product cofactor S-adenosylhomocysteine. Histone H4 intercalates in the substrate-binding cleft as an extended parallel beta-strand. Residues preceding Lys-20 in H4 engage in an extensive array of salt bridge, hydrogen bond, and van der Waals interactions with hSET8, while the C-terminal residues bind through predominantly hydrophobic interactions. Mutational analysis of both the substrate-binding cleft and histone H4 reveals that interactions with residues in the N and C termini of the H4 peptide are critical for conferring substrate specificity. Finally, analysis of the product specificity indicates that hSET8 is a monomethylase, consistent with its role in the maintenance of Lys-20 monomethylation during cell division.
Figures


References
-
- Aasland R., Abrams, C., Ampe, C., Ball, L.J., Bedford, M.T., Cesareni, G., Gimona, M., Hurley, J.H., Jarchau, T., Lehto, V.P., et al. 2002. Normalization of nomenclature for peptide motifs as ligands of modular protein domains. FEBS Lett. 513: 141-144. - PubMed
-
- Abrahams J.P. and Leslie, A.G.W. 1996. Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. D Biol. Crystallogr. 52: 30-42. - PubMed
-
- Beisel C., Imhof, A., Greene, J., Kremmer, E., and Sauer, F. 2002. Histone methylation by the Drosophila epigenetic transcriptional regulator Ash1. Nature 419: 857-862. - PubMed
-
- Brunzelle J.S., Shafaee, P., Yang, X., Weigand, S., Ren, Z., and Anderson, W.F. 2003. Automated crystallographic system for high-throughput protein structure determination. Acta Crystallogr. D Biol. Crystallogr. 59: 1138-1144. - PubMed
-
- Chirpich T.P., Zappia, V., Costilow, R.N., and Barker, H.A. 1970. Lysine 2,3-aminomutase. Purification and properties of a pyridoxal phosphate and S-adenosylmethionine-activated enzyme. J. Biol. Chem. 245: 1778-1789. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
