Structural basis for recruitment of RILP by small GTPase Rab7
- PMID: 15933719
- PMCID: PMC1142575
- DOI: 10.1038/sj.emboj.7600643
Structural basis for recruitment of RILP by small GTPase Rab7
Abstract
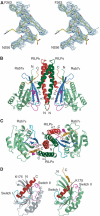
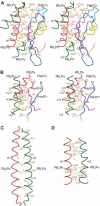
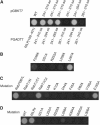
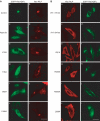
Rab7 regulates vesicle traffic from early to late endosomes, and from late endosomes to lysosomes. The crystal structure of Rab7-GTP in complex with the Rab7 binding domain of RILP reveals that Rab7 interacts with RILP specifically via two distinct areas, with the first one involving the switch and interswitch regions and the second one consisting of RabSF1 and RabSF4. Disruption of these interactions by mutations abrogates late endosomal/lysosomal targeting of Rab7 and RILP. The Rab7 binding domain of RILP forms a coiled-coil homodimer with two symmetric surfaces to interact with two separate Rab7-GTP molecules, forming a dyad configuration of Rab7-RILP(2)-Rab7. Mutations that disrupt RILP dimerization also abolish its interactions with Rab7-GTP and late endosomal/lysosomal targeting, suggesting that the dimeric form of RILP is a functional unit. Structural comparison suggests that the combined use of RabSF1 and RabSF4 with the switch regions may be a general mode of action for most Rab proteins in regulating membrane trafficking.
Figures


Similar articles
-
The Rab-interacting lysosomal protein, a Rab7 and Rab34 effector, is capable of self-interaction.Biochem Biophys Res Commun. 2005 Aug 19;334(1):128-33. doi: 10.1016/j.bbrc.2005.06.067. Biochem Biophys Res Commun. 2005. PMID: 15996637
-
Rab-interacting lysosomal protein (RILP): the Rab7 effector required for transport to lysosomes.EMBO J. 2001 Feb 15;20(4):683-93. doi: 10.1093/emboj/20.4.683. EMBO J. 2001. PMID: 11179213 Free PMC article.
-
Structural basis of human ORP1-Rab7 interaction for the late-endosome and lysosome targeting.PLoS One. 2019 Feb 5;14(2):e0211724. doi: 10.1371/journal.pone.0211724. eCollection 2019. PLoS One. 2019. PMID: 30721249 Free PMC article.
-
Rab7: role of its protein interaction cascades in endo-lysosomal traffic.Cell Signal. 2011 Mar;23(3):516-21. doi: 10.1016/j.cellsig.2010.09.012. Epub 2010 Sep 21. Cell Signal. 2011. PMID: 20851765 Review.
-
Rab7 and the CMT2B disease.Biochem Soc Trans. 2009 Oct;37(Pt 5):1027-31. doi: 10.1042/BST0371027. Biochem Soc Trans. 2009. PMID: 19754445 Review.
Cited by
-
Crystal structure of the FYCO1 RUN domain suggests possible interfaces with small GTPases.Acta Crystallogr F Struct Biol Commun. 2020 Aug 1;76(Pt 8):326-333. doi: 10.1107/S2053230X20009012. Epub 2020 Jul 28. Acta Crystallogr F Struct Biol Commun. 2020. PMID: 32744243 Free PMC article.
-
USP32 regulates late endosomal transport and recycling through deubiquitylation of Rab7.Nat Commun. 2019 Mar 29;10(1):1454. doi: 10.1038/s41467-019-09437-x. Nat Commun. 2019. PMID: 30926795 Free PMC article.
-
The structural basis of Arf effector specificity: the crystal structure of ARF6 in a complex with JIP4.EMBO J. 2009 Sep 16;28(18):2835-45. doi: 10.1038/emboj.2009.209. Epub 2009 Jul 30. EMBO J. 2009. PMID: 19644450 Free PMC article.
-
Charcot Marie Tooth 2B Peripheral Sensory Neuropathy: How Rab7 Mutations Impact NGF Signaling?Int J Mol Sci. 2017 Feb 4;18(2):324. doi: 10.3390/ijms18020324. Int J Mol Sci. 2017. PMID: 28165391 Free PMC article. Review.
-
Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking.Nat Cell Biol. 2008 Jul;10(7):776-87. doi: 10.1038/ncb1740. Epub 2008 Jun 15. Nat Cell Biol. 2008. PMID: 18552835 Free PMC article.
References
-
- Ali BR, Wasmeier C, Lamoreux L, Strom M, Seabra MC (2004) Multiple regions contribute to membrane targeting of Rab GTPases. J Cell Sci 117: 6401–6412 - PubMed
-
- Andres DA, Seabra MC, Brown MS, Armstrong SA, Smeland TE, Cremers FP, Goldstein JL (1993) cDNA cloning of component A of Rab geranylgeranyl transferase and demonstration of its role as a Rab escort protein. Cell 73: 1091–1099 - PubMed
-
- Brennwald P, Novick P (1993) Interactions of three domains distinguishing the Ras-related GTP-binding proteins Ypt1 and Sec4. Nature 362: 560–563 - PubMed
-
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D 54: 905–921 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

