Translation of aberrant mRNAs lacking a termination codon or with a shortened 3'-UTR is repressed after initiation in yeast
- PMID: 15933721
- PMCID: PMC1142571
- DOI: 10.1038/sj.emboj.7600636
Translation of aberrant mRNAs lacking a termination codon or with a shortened 3'-UTR is repressed after initiation in yeast
Abstract
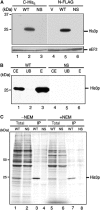
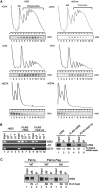
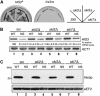
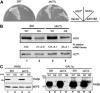
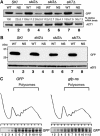
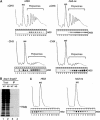
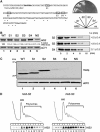
A novel mRNA surveillance for mRNA lacking a termination codon (nonstop mRNA) has been proposed in which Ski7p is thought to recognize stalled ribosomes at the 3' end of mRNA. Here we report our analysis of translation and decay of nonstop mRNAs in Saccharomyces cerevisiae. Although the reduction of nonstop mRNAs was only 4.5-fold, a level that is sufficient for residual protein synthesis, translation products of nonstop mRNAs were hardly detectable. We show that nonstop mRNAs were associated with polysomes, but not with Pab1p. We also show that ribosomes translating nonstop mRNA formed stable and heavy polysome complexes with mRNA. These data suggest that ribosome stalling at the 3' end of nonstop mRNA may block further rounds of translation, hence repressing protein synthesis. Furthermore, it was found that the 5' --> 3' decay pathway was accelerated for nonstop mRNA decay in the absence of Ski7p. We also found that translation of aberrant mRNAs with a shortened 3'-UTR was repressed, suggesting that an improper spatial distance between the termination codon and the 3' end of mRNA results in translation repression.
Figures



References
-
- Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A (2004) A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432: 112–118 - PubMed
-
- Caponigro G, Parker R (1995) Multiple functions for the poly(A)-binding protein in mRNA decapping and deadenylation in yeast. Genes Dev 9: 2421–2432 - PubMed
-
- Coller J, Parker R (2004) Eukaryotic mRNA decapping. Annu Rev Biochem 73: 861–890 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

