Nova autoregulation reveals dual functions in neuronal splicing
- PMID: 15933722
- PMCID: PMC1142566
- DOI: 10.1038/sj.emboj.7600630
Nova autoregulation reveals dual functions in neuronal splicing
Abstract
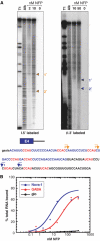
The Nova family of neuron-specific RNA-binding proteins were originally identified as targets in an autoimmune neurologic disease characterized by failure of motor inhibition. Nova-1 regulates alternative splicing of pre-mRNAs encoding the inhibitory neurotransmitter receptor subunits GABA(A)Rgamma2 and GlyRalpha2 by directly binding intronic elements, resulting in enhancement of exon inclusion. Here we identify exon E4 in the Nova-1 pre-mRNA itself, encoding a phosphorylated protein domain, as an additional target of Nova-dependent splicing regulation in the mouse spinal cord. Nova binding to E4 is necessary and sufficient for Nova-dependent exon exclusion. E4 harbors five repeats of the known Nova-binding tetranucleotide YCAY and mutation of these elements destroys Nova-dependent regulation. Furthermore, swapping of the sites from Nova-1 and GABA(A)Rgamma2 indicates that the ability of Nova to enhance or repress alternative exon inclusion is dependent on the position of the Nova-binding element within the pre-mRNA. These studies demonstrate that in addition to its previously described role as a splicing activator, Nova autoregulates its own expression by acting as a splicing repressor.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

