Extensive de Novo genomic variation in rice induced by introgression from wild rice (Zizania latifolia Griseb.)
- PMID: 15937131
- PMCID: PMC1449789
- DOI: 10.1534/genetics.105.040964
Extensive de Novo genomic variation in rice induced by introgression from wild rice (Zizania latifolia Griseb.)
Abstract
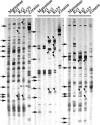
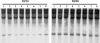
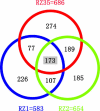
To study the possible impact of alien introgression on a recipient plant genome, we examined >6000 unbiased genomic loci of three stable rice recombinant inbred lines (RILs) derived from intergeneric hybridization between rice (cv. Matsumae) and a wild relative (Zizania latifolia Griseb.) followed by successive selfing. Results from amplified fragment length polymorphism (AFLP) analysis showed that, whereas the introgressed Zizania DNA comprised <0.1% of the genome content in the RILs, extensive and genome-wide de novo variations occurred in up to 30% of the analyzed loci for all three lines studied. The AFLP-detected changes were validated by DNA gel-blot hybridization and/or sequence analysis of genomic loci corresponding to a subset of the differentiating AFLP fragments. A BLAST analysis revealed that the genomic variations occurred in diverse sequences, including protein-coding genes, transposable elements, and sequences of unknown functions. Pairwise sequence comparison of selected loci between a RIL and its rice parent showed that the variations represented either base substitutions or small insertion/deletions. Genome variations were detected in all 12 rice chromosomes, although their distribution was uneven both among and within chromosomes. Taken together, our results imply that even cryptic alien introgression can be highly mutagenic to a recipient plant genome.
Figures




Similar articles
-
Extensive microsatellite variation in rice induced by introgression from wild rice (Zizania latifolia Griseb.).PLoS One. 2013 Apr 24;8(4):e62317. doi: 10.1371/journal.pone.0062317. Print 2013. PLoS One. 2013. PMID: 23638037 Free PMC article.
-
Extent and pattern of DNA methylation alteration in rice lines derived from introgressive hybridization of rice and Zizania latifolia Griseb.Theor Appl Genet. 2006 Jul;113(2):196-205. doi: 10.1007/s00122-006-0286-2. Epub 2006 May 5. Theor Appl Genet. 2006. PMID: 16791687
-
Extensive alterations in DNA methylation and transcription in rice caused by introgression from Zizania latifolia.Plant Mol Biol. 2004 Mar;54(4):571-82. doi: 10.1023/B:PLAN.0000038270.48326.7a. Plant Mol Biol. 2004. PMID: 15316290
-
Alien introgression in rice.Plant Mol Biol. 1997 Sep;35(1-2):35-47. Plant Mol Biol. 1997. PMID: 9291958 Review.
-
The genetic colinearity of rice and other cereals on the basis of genomic sequence analysis.Curr Opin Plant Biol. 2003 Apr;6(2):128-33. doi: 10.1016/s1369-5266(03)00015-3. Curr Opin Plant Biol. 2003. PMID: 12667868 Review.
Cited by
-
Contrasting patterns of genetic and phenotypic divergence of two sympatric congeners, Phragmites australis and P. hirsuta, in heterogeneous habitats.Front Plant Sci. 2023 Dec 12;14:1299128. doi: 10.3389/fpls.2023.1299128. eCollection 2023. Front Plant Sci. 2023. PMID: 38162310 Free PMC article.
-
Linkage maps of the dwarf and Normal lake whitefish (Coregonus clupeaformis) species complex and their hybrids reveal the genetic architecture of population divergence.Genetics. 2007 Jan;175(1):375-98. doi: 10.1534/genetics.106.061457. Epub 2006 Nov 16. Genetics. 2007. PMID: 17110497 Free PMC article.
-
Transgenerational activation of an autonomous DNA transposon, Dart1-24, by 5-azaC treatment in rice.Theor Appl Genet. 2019 Dec;132(12):3347-3355. doi: 10.1007/s00122-019-03429-7. Epub 2019 Oct 3. Theor Appl Genet. 2019. PMID: 31583438
-
Mobile DNA and evolution in the 21st century.Mob DNA. 2010 Jan 25;1(1):4. doi: 10.1186/1759-8753-1-4. Mob DNA. 2010. PMID: 20226073 Free PMC article.
-
Reconstituting the genome of a young allopolyploid crop, Brassica napus, with its related species.Plant Biotechnol J. 2019 Jun;17(6):1106-1118. doi: 10.1111/pbi.13041. Epub 2019 Jan 7. Plant Biotechnol J. 2019. PMID: 30467941 Free PMC article.
References
-
- Adams, K. L., and J. F. Wendel, 2004. Exploring the genomic mysteries of polyploidy in cotton. Biol. J. Linn. Soc. 82: 573–581.
-
- Adams, K. L., and J. F. Wendel, 2005. Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 8: 135–141. - PubMed
-
- Anderson, E., 1949 Introgressive Hybridization. John Wiley & Sons, New York.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

