Temperature-dependent conjugative transfer of R27: role of chromosome- and plasmid-encoded Hha and H-NS proteins
- PMID: 15937157
- PMCID: PMC1151748
- DOI: 10.1128/JB.187.12.3950-3959.2005
Temperature-dependent conjugative transfer of R27: role of chromosome- and plasmid-encoded Hha and H-NS proteins
Abstract
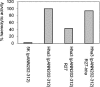
IncHI plasmids encode multiple-antibiotic resistance in Salmonella enterica serovar Typhi. These plasmids have been considered to play a relevant role in the persistence and reemergence of this microorganism. The IncHI1 plasmid R27, which can be considered the prototype of IncHI plasmids, is thermosensitive for transfer. Conjugation frequency is highest at low temperature (25 to 30 degrees C), decreasing when temperature increases. R27 codifies an H-NS-like protein (open reading frame 164 [ORF164]) and an Hha-like protein (ORF182). The H-NS and Hha proteins participate in the thermoregulation of gene expression in Escherichia coli. Here we investigated the hypothetical role of such proteins in thermoregulation of R27 conjugation. At a nonpermissive temperature (33 degrees C), transcription of several ORFs in both transfer region 1 (Tra1) and Tra2 from R27 is upregulated in cells depleted of Hha-like and H-NS-like proteins. Both chromosome- and plasmid-encoded Hha and H-NS proteins appear to potentially modulate R27 transfer. The function of R27-encoded Hha-like and H-NS proteins is not restricted to modulation of R27 transfer. Different mutant phenotypes associated with both chromosomal hha and hns mutations are compensated in cells harboring R27.
Figures





References
-
- Atlung, T., and H. Ingmer. 1997. H-NS: a modulator of environmentally regulated gene expression. Mol. Microbiol. 24:7-17. - PubMed
-
- Beloin, C., P. Deighan, M. Doyle, and C. J. Dorman. 2003. Shigella flexneri 2a strain 2457T expresses three members of the H-NS-like protein family: characterization of the Sfh protein. Mol. Gen. Genomics 270:66-77. - PubMed
-
- Bloch, V., Y. Yang, E. Margeat, A. Chavanieu, M. T. Augé, B. Robert, S. Arold, S. Rimsky, and M. Kochoyan. 2003. The H-NS dimerization domain defines a new fold contributing to DNA recognition. Nat. Struct. Biol. 10:212-218. - PubMed
-
- Cornelis, G. R., C. Sluiters, I. Delor, D. Gelb, K. Kaninga, C. Lambert de Rouvroit, M.-P. Sory, J.-C. Vanooteghem, and T. Michiels. 1991. ymoA, a Yersinia enterocolitica chromosomal gene modulating the expression of virulence functions. Mol. Microbiol. 5:1023-1034. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

