Regulation and function of cytochrome c' in Rhodobacter sphaeroides 2.4.3
- PMID: 15937170
- PMCID: PMC1151734
- DOI: 10.1128/JB.187.12.4077-4085.2005
Regulation and function of cytochrome c' in Rhodobacter sphaeroides 2.4.3
Abstract
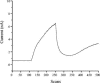
Cytochrome c' (Cyt c') is a c-type cytochrome with a pentacoordinate heme iron. The gene encoding this protein in Rhodobacter sphaeroides 2.4.3, designated cycP, was isolated and sequenced. Northern blot analysis and beta-galactosidase assays demonstrated that cycP transcription increased as oxygen levels decreased and was not repressed under denitrifying conditions as observed in another Rhodobacter species. CO difference spectra performed with extracts of cells grown under different conditions revealed that Cyt c' levels were highest during photosynthetic denitrifying growth conditions. The increase in Cyt c' under this condition was higher than would be predicted from transcriptional studies. Electron paramagnetic resonance analysis of whole cells demonstrated that Cyt c' binds NO during denitrification. Mass spectrometric analysis of nitrogen oxides produced by cells and purified protein did not indicate that Cyt c' has NO reductase activity. Taken together, these results suggest a model where Cyt c' in R. sphaeroides 2.4.3 may shuttle NO to the membrane, where it can be reduced.
Figures


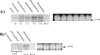



References
-
- Andrew, C. R., S. J. George, D. M. Lawson, and R. R. Eady. 2002. Six- to five-coordinate heme-nitrosyl conversion in cytochrome c′ and its relevance to guanylate cyclase. Biochemistry 41:2353-2360. - PubMed
-
- Bartnikas, T. B., Y. Wang, T. Bobo, A. Veselov, C. P. Scholes, and J. P. Shapleigh. 2002. Characterization of a member of the NnrR regulon in Rhodobacter sphaeroides 2.4.3 encoding a haem-copper protein. Microbiology 148:825-833. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

