Characterization of Gla(KP), a UDP-galacturonic acid C4-epimerase from Klebsiella pneumoniae with extended substrate specificity
- PMID: 15937173
- PMCID: PMC1151718
- DOI: 10.1128/JB.187.12.4104-4115.2005
Characterization of Gla(KP), a UDP-galacturonic acid C4-epimerase from Klebsiella pneumoniae with extended substrate specificity
Abstract
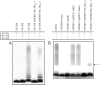
In Escherichia coli and Salmonella enterica, the core oligosaccharide backbone of the lipopolysaccharide is modified by phosphoryl groups. The negative charges provided by these residues are important in maintaining the barrier function of the outer membrane. In contrast, Klebsiella pneumoniae lacks phosphoryl groups in its core oligosaccharide but instead contains galacturonic acid residues that are proposed to serve a similar function in outer membrane stability. Gla(KP) is a UDP-galacturonic acid C4-epimerase that provides UDP-galacturonic acid for core synthesis, and the enzyme was biochemically characterized because of its potentially important role in outer membrane stability. High-performance anion-exchange chromatography was used to demonstrate the UDP-galacturonic acid C4-epimerase activity of Gla(KP), and capillary electrophoresis was used for activity assays. The reaction equilibrium favors UDP-galacturonic acid over UDP-glucuronic acid in a ratio of 1.4:1, with the K(m) for UDP-glucuronic acid of 13.0 microM. Gla(KP) exists as a dimer in its native form. NAD+/NADH is tightly bound by the enzyme and addition of supplementary NAD+ is not required for activity of the purified enzyme. Divalent cations have an unexpected inhibitory effect on enzyme activity. Gla(KP) was found to have a broad substrate specificity in vitro; it is capable of interconverting UDP-glucose/UDP-galactose and UDP-N-acetylglucosamine/UDP-N-acetylgalactosamine, albeit at much lower activity. The epimerase GalE interconverts UDP-glucose/UDP-galactose. Multicopy plasmid-encoded gla(KP) partially complemented a galE mutation in S. enterica and in K. pneumoniae; however, chromosomal gla(KP) could not substitute for galE in a K. pneumoniae galE mutant in vivo.
Figures




References
-
- Bagley, S. T. 1985. Habitat association of Klebsiella species. Infect. Control 6:52-58. - PubMed
-
- Bauer, A. J., I. Rayment, P. A. Frey, and H. M. Holden. 1992. The molecular structure of UDP-galactose 4-epimerase from Escherichia coli determined at 2.5 Å resolution. Proteins 12:372-381. - PubMed
-
- Bengoechea, J. A., E. Pinta, T. Salminen, C. Oertelt, O. Holst, J. Radziejewska-Lebrecht, Z. Piotrowska-Seget, R. Venho, and M. Skurnik. 2002. Functional characterization of Gne (UDP-N-acetylglucosamine-4-epimerase), Wzz (chain length determinant), and Wzy (O-antigen polymerase) of Yersinia enterocolitica serotype O:8. J. Bacteriol. 184:4277-4287. - PMC - PubMed
-
- Bernatchez, S., C. M. Szymanski, N. Ishiyama, J. Li, H. C. Jarrell, P. C. Lau, A. M. Berghuis, N. M. Young, and W. W. Wakarchuk. 2004. A single bifunctional UDP-GlcNAc/Glc 4-epimerase supports the synthesis of three cell surface glycoconjugates in Campylobacter jejuni. J. Biol. Chem. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

