Structural characterization and assembly of the distal tail structure of the temperate lactococcal bacteriophage TP901-1
- PMID: 15937180
- PMCID: PMC1151708
- DOI: 10.1128/JB.187.12.4187-4197.2005
Structural characterization and assembly of the distal tail structure of the temperate lactococcal bacteriophage TP901-1
Abstract
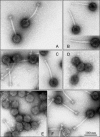
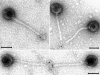
The tail structures of bacteriophages infecting gram-positive bacteria are largely unexplored, although the phage tail mediates the initial interaction with the host cell. The temperate Lactococcus lactis phage TP901-1 of the Siphoviridae family has a long noncontractile tail with a distal baseplate. In the present study, we investigated the distal tail structures and tail assembly of phage TP901-1 by introducing nonsense mutations into the late transcribed genes dit (orf46), tal(TP901-1) (orf47), bppU (orf48), bppL (orf49), and orf50. Transmission electron microscopy examination of mutant and wild-type TP901-1 phages showed that the baseplate consisted of two different disks and that a central tail fiber is protruding below the baseplate. Evaluation of the mutant tail morphologies with protein profiles and Western blots revealed that the upper and lower baseplate disks consist of the proteins BppU and BppL, respectively. Likewise, Dit and Tal(TP901-1) were shown to be structural tail proteins essential for tail formation, and Tal(TP901-1) was furthermore identified as the tail fiber protein by immunogold labeling experiments. Determination of infection efficiencies of the mutant phages showed that the baseplate is fundamental for host infection and the lower disk protein, BppL, is suggested to interact with the host receptor. In contrast, ORF50 was found to be nonessential for tail assembly and host infection. A model for TP901-1 tail assembly, in which the function of eight specific proteins is considered, is presented.
Figures



Similar articles
-
Identification of the lower baseplate protein as the antireceptor of the temperate lactococcal bacteriophages TP901-1 and Tuc2009.J Bacteriol. 2006 Jan;188(1):55-63. doi: 10.1128/JB.188.1.55-63.2006. J Bacteriol. 2006. PMID: 16352821 Free PMC article.
-
Mutational analysis of two structural genes of the temperate lactococcal bacteriophage TP901-1 involved in tail length determination and baseplate assembly.Virology. 2000 Oct 25;276(2):315-28. doi: 10.1006/viro.2000.0497. Virology. 2000. PMID: 11040123
-
The Atomic Structure of the Phage Tuc2009 Baseplate Tripod Suggests that Host Recognition Involves Two Different Carbohydrate Binding Modules.mBio. 2016 Jan 26;7(1):e01781-15. doi: 10.1128/mBio.01781-15. mBio. 2016. PMID: 26814179 Free PMC article.
-
Conserved and Diverse Traits of Adhesion Devices from Siphoviridae Recognizing Proteinaceous or Saccharidic Receptors.Viruses. 2020 May 6;12(5):512. doi: 10.3390/v12050512. Viruses. 2020. PMID: 32384698 Free PMC article. Review.
-
Structures and host-adhesion mechanisms of lactococcal siphophages.Front Microbiol. 2014 Jan 16;5:3. doi: 10.3389/fmicb.2014.00003. eCollection 2014. Front Microbiol. 2014. PMID: 24474948 Free PMC article. Review.
Cited by
-
A Novel Genus of Actinobacterial Tectiviridae.Viruses. 2019 Dec 7;11(12):1134. doi: 10.3390/v11121134. Viruses. 2019. PMID: 31817897 Free PMC article.
-
Phages of non-dairy lactococci: isolation and characterization of ΦL47, a phage infecting the grass isolate Lactococcus lactis ssp. cremoris DPC6860.Front Microbiol. 2014 Jan 13;4:417. doi: 10.3389/fmicb.2013.00417. eCollection 2014 Jan 13. Front Microbiol. 2014. PMID: 24454309 Free PMC article.
-
Morphology, genome sequence, and structural proteome of type phage P335 from Lactococcus lactis.Appl Environ Microbiol. 2008 Aug;74(15):4636-44. doi: 10.1128/AEM.00118-08. Epub 2008 Jun 6. Appl Environ Microbiol. 2008. PMID: 18539805 Free PMC article.
-
A shifty stop for a hairy tail.Mol Microbiol. 2008 Nov;70(3):549-53. doi: 10.1111/j.1365-2958.2008.06434.x. Epub 2008 Sep 12. Mol Microbiol. 2008. PMID: 18826406 Free PMC article.
-
The lactococcal phages Tuc2009 and TP901-1 incorporate two alternate forms of their tail fiber into their virions for infection specialization.J Biol Chem. 2013 Feb 22;288(8):5581-90. doi: 10.1074/jbc.M112.444901. Epub 2013 Jan 8. J Biol Chem. 2013. PMID: 23300085 Free PMC article.
References
-
- Blatny, J. M., L. Godager, M. Lunde, and I. F. Nes. 2004. Complete genome sequence of the Lactococcus lactis temperate phage ϕLC3: comparative analysis of ϕLC3 and its relatives in lactococci and streptococci. Virology 318:231-244. - PubMed
-
- Böhm, J., O. Lambert, A. S. Frangakis, L. Letellier, W. Baumeister, and J. L. Rigaud. 2001. FhuA-mediated phage genome transfer into liposomes: a cryo-electron tomography study. Curr. Biol. 11:1168-1175. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

