Transcriptional profile of the Arabidopsis root quiescent center
- PMID: 15937229
- PMCID: PMC1167541
- DOI: 10.1105/tpc.105.031724
Transcriptional profile of the Arabidopsis root quiescent center
Abstract
The self-renewal characteristics of stem cells render them vital engines of development. To better understand the molecular mechanisms that determine the properties of stem cells, transcript profiling was conducted on quiescent center (QC) cells from the Arabidopsis thaliana root meristem. The AGAMOUS-LIKE 42 (AGL42) gene, which encodes a MADS box transcription factor whose expression is enriched in the QC, was used to mark these cells. RNA was isolated from sorted cells, labeled, and hybridized to Affymetrix microarrays. Comparisons with digital in situ expression profiles of surrounding tissues identified a set of genes enriched in the QC. Promoter regions from a subset of transcription factors identified as enriched in the QC conferred expression in the QC. These studies demonstrated that it is possible to successfully isolate and profile a rare cell type in the plant. Mutations in all enriched transcription factor genes including AGL42 exhibited no detectable root phenotype, raising the possibility of a high degree of functional redundancy in the QC.
Figures


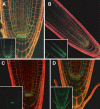


References
-
- Aida, M., Beis, D., Heidstra, R., Willemsen, V., Blilou, I., Galinha, C., Nussaume, L., Noh, Y.S., Amasino, R., and Scheres, B. (2004). The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119, 109–120. - PubMed
-
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. - PubMed
-
- Aoyama, T., and Chua, N.H. (1997). A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11, 605–612. - PubMed
-
- Beissbarth, T., and Speed, T.P. (2004). GOstat: Find statistically overrepresented gene ontologies within a group of genes. Bioinformatics 20, 1464–1465. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

