Assessing strategies for improved superfamily recognition
- PMID: 15937274
- PMCID: PMC2253352
- DOI: 10.1110/ps.041056105
Assessing strategies for improved superfamily recognition
Abstract
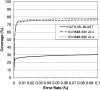
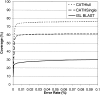
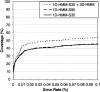
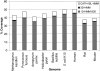
There are more than 200 completed genomes and over 1 million nonredundant sequences in public repositories. Although the structural data are more sparse (approximately 13,000 nonredundant structures solved to date), several powerful sequence-based methodologies now allow these structures to be mapped onto related regions in a significant proportion of genome sequences. We review a number of publicly available strategies for providing structural annotations for genome sequences, and we describe the protocol adopted to provide CATH structural annotations for completed genomes. In particular, we assess the performance of several sequence-based protocols employing Hidden Markov model (HMM) technologies for superfamily recognition, including a new approach (SAMOSA [sequence augmented models of structure alignments]) that exploits multiple structural alignments from the CATH domain structure database when building the models. Using a data set of remote homologs detected by structure comparison and manually validated in CATH, a single-seed HMM library was able to recognize 76% of the data set. Including the SAMOSA models in the HMM library showed little gain in homolog recognition, although a slight improvement in alignment quality was observed for very remote homologs. However, using an expanded 1D-HMM library, CATH-ISL increased the coverage to 86%. The single-seed HMM library has been used to annotate the protein sequences of 120 genomes from all three major kingdoms, allowing up to 70% of the genes or partial genes to be assigned to CATH superfamilies. It has also been used to recruit sequences from Swiss-Prot and TrEMBL into CATH domain superfamilies, expanding the CATH database eightfold.
Figures




References
-
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215 403–410. - PubMed
-
- Attwood, T.,K., Avison, H., Beck, M.E., Bewley, M., Bleasby, A.J., Brewster, F., Cooper, P., Degtyarenko, K., Geddes, A.J., Flower, D.R., et al. 1997. The PRINTS database of protein fingerprints: A novel information resource for computational molecular biology. J. Chem. Inf. Comput. Sci. 37 417–424. - PubMed
-
- Barton, G. and Sternberg, M. 1987. A strategy for the rapid multiple alignment of protein sequences. Confidence levels from tertiary structure comparisons. J. Mol. Biol. 198 327–337. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

