Crystal structure of methylenetetrahydromethanopterin reductase (Mer) in complex with coenzyme F420: Architecture of the F420/FMN binding site of enzymes within the nonprolyl cis-peptide containing bacterial luciferase family
- PMID: 15937276
- PMCID: PMC2253363
- DOI: 10.1110/ps.041289805
Crystal structure of methylenetetrahydromethanopterin reductase (Mer) in complex with coenzyme F420: Architecture of the F420/FMN binding site of enzymes within the nonprolyl cis-peptide containing bacterial luciferase family
Abstract
Methylenetetratetrahydromethanopterin reductase (Mer) is involved in CO(2) reduction to methane in methanogenic archaea and catalyses the reversible reduction of methylenetetrahydromethanopterin (methylene-H(4)MPT) to methyl-H(4)MPT with coenzyme F(420)H(2), which is a reduced 5'-deazaflavin. Mer was recently established as a TIM barrel structure containing a nonprolyl cis-peptide bond but the binding site of the substrates remained elusive. We report here on the crystal structure of Mer in complex with F(420) at 2.6 A resolution. The isoalloxazine ring is present in a pronounced butterfly conformation, being induced from the Re-face of F(420) by a bulge that contains the non-prolyl cis-peptide bond. The bindingmode of F(420) is very similar to that in F(420)-dependent alcohol dehydrogenase Adf despite the low sequence identity of 21%. Moreover, binding of F(420) to the apoenzyme was only associated with minor conformational changes of the polypeptide chain. These findings allowed us to build an improved model of FMN into its binding site in bacterial luciferase, which belongs to the same structural family as Mer and Adf and also contains a nonprolyl cis-peptide bond in an equivalent position.
Figures

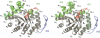

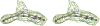
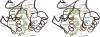
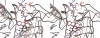

Similar articles
-
Structure of coenzyme F(420) dependent methylenetetrahydromethanopterin reductase from two methanogenic archaea.J Mol Biol. 2000 Jul 21;300(4):935-50. doi: 10.1006/jmbi.2000.3909. J Mol Biol. 2000. PMID: 10891279
-
Coenzyme binding in F420-dependent secondary alcohol dehydrogenase, a member of the bacterial luciferase family.Structure. 2004 Mar;12(3):361-70. doi: 10.1016/j.str.2004.02.010. Structure. 2004. PMID: 15016352
-
Coenzyme F420-dependent N5,N10-methylenetetrahydromethanopterin reductase (Mer) from Methanobacterium thermoautotrophicum strain Marburg. Cloning, sequencing, transcriptional analysis, and functional expression in Escherichia coli of the mer gene.Eur J Biochem. 1995 Aug 1;231(3):773-8. doi: 10.1111/j.1432-1033.1995.0773d.x. Eur J Biochem. 1995. PMID: 7649177
-
Re-face specificity at C14a of methylenetetrahydromethanopterin and Si-face specificity at C5 of coenzyme F420 for coenzyme F420-dependent methylenetetrahydromethanopterin dehydrogenase from methanogenic Archaea.Eur J Biochem. 1995 Jan 15;227(1-2):169-74. doi: 10.1111/j.1432-1033.1995.tb20373.x. Eur J Biochem. 1995. PMID: 7851382
-
Crystal structure of DMGO provides a prototype for a new tetrahydrofolate-binding fold.Biochem Soc Trans. 2005 Aug;33(Pt 4):776-9. doi: 10.1042/BST0330776. Biochem Soc Trans. 2005. PMID: 16042597 Review.
Cited by
-
A previously undescribed pathway for pyrimidine catabolism.Proc Natl Acad Sci U S A. 2006 Mar 28;103(13):5114-9. doi: 10.1073/pnas.0600521103. Epub 2006 Mar 15. Proc Natl Acad Sci U S A. 2006. PMID: 16540542 Free PMC article.
-
Molecular insights into the biosynthesis of the F420 coenzyme.J Biol Chem. 2008 Apr 25;283(17):11832-40. doi: 10.1074/jbc.M710352200. Epub 2008 Feb 5. J Biol Chem. 2008. PMID: 18252724 Free PMC article.
-
F420H2 Is Required for Phthiocerol Dimycocerosate Synthesis in Mycobacteria.J Bacteriol. 2016 Jul 13;198(15):2020-8. doi: 10.1128/JB.01035-15. Print 2016 Aug 1. J Bacteriol. 2016. PMID: 27185825 Free PMC article.
-
Cofactor F420: an expanded view of its distribution, biosynthesis and roles in bacteria and archaea.FEMS Microbiol Rev. 2021 Sep 8;45(5):fuab021. doi: 10.1093/femsre/fuab021. FEMS Microbiol Rev. 2021. PMID: 33851978 Free PMC article.
-
A Novel F420-dependent Thioredoxin Reductase Gated by Low Potential FAD: A TOOL FOR REDOX REGULATION IN AN ANAEROBE.J Biol Chem. 2016 Oct 28;291(44):23084-23100. doi: 10.1074/jbc.M116.750208. Epub 2016 Sep 2. J Biol Chem. 2016. PMID: 27590343 Free PMC article.
References
-
- Abu-Soud, H.M., Clark, A.C., Francisco, W.A., Baldwin, T.O., and Raushel, F.M. 1993. Kinetic destabilization of the hydroperoxy flavin intermediate by site-directed modification of the reactive thiol in bacterial luciferase. J. Biol. Chem. 268 7699–7706. - PubMed
-
- Acharya, P., Goenrich, M., Hagemeier, C.H., Demmer, U., Vorholt, J., Thauer, R.K., and Ermler, U. 2005. How an enzyme binds the C1-carrier tetrahydromethanopterin: Structure of the tetrahydromethanopterin dependent formaldehyde-activating enzyme Fae from Methylobacterium extorquens AM1. J. Biol. Chem. (in press). - PubMed
-
- Ahn, H.J., Yoon, H.J., Lee 2d, B., and Suh, S.W. 2004. Crystal structure of chorismate synthase: A novel FMN-binding protein fold and functional insights. J. Mol. Biol. 336 903–915. - PubMed
-
- Aufhammer, S.W., Warkentin, E., Berk, H., Shima, S., Thauer, R.K., and Ermler, U. 2004. Coenzyme binding in F420-dependent secondary alcohol dehydrogenase, a member of the bacterial luciferase family. Structure 12 361–370. - PubMed
-
- Bacon, D.J. and Anderson, W.F. 1988. A fast algorithm for rendering space-filling molecule pictures. J. Mol. Graph. 6 219–220.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

