The individual and common repertoire of DNA-binding transcriptional regulators of Corynebacterium glutamicum, Corynebacterium efficiens, Corynebacterium diphtheriae and Corynebacterium jeikeium deduced from the complete genome sequences
- PMID: 15938759
- PMCID: PMC1180825
- DOI: 10.1186/1471-2164-6-86
The individual and common repertoire of DNA-binding transcriptional regulators of Corynebacterium glutamicum, Corynebacterium efficiens, Corynebacterium diphtheriae and Corynebacterium jeikeium deduced from the complete genome sequences
Abstract
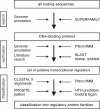
Background: The genus Corynebacterium includes Gram-positive microorganisms of great biotechnologically importance, such as Corynebacterium glutamicum and Corynebacterium efficiens, as well as serious human pathogens, such as Corynebacterium diphtheriae and Corynebacterium jeikeium. Although genome sequences of the respective species have been determined recently, the knowledge about the repertoire of transcriptional regulators and the architecture of global regulatory networks is scarce. Here, we apply a combination of bioinformatic tools and a comparative genomic approach to identify and characterize a set of conserved DNA-binding transcriptional regulators in the four corynebacterial genomes.
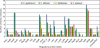
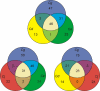
Results: A collection of 127 DNA-binding transcriptional regulators was identified in the C. glutamicum ATCC 13032 genome, whereas 103 regulators were detected in C. efficiens YS-314, 63 in C. diphtheriae NCTC 13129 and 55 in C. jeikeium K411. According to amino acid sequence similarities and protein structure predictions, the DNA-binding transcriptional regulators were grouped into 25 regulatory protein families. The common set of DNA-binding transcriptional regulators present in the four corynebacterial genomes consists of 28 proteins that are apparently involved in the regulation of cell division and septation, SOS and stress response, carbohydrate metabolism and macroelement and metal homeostasis.
Conclusion: This work describes characteristic features of a set of conserved DNA-binding transcriptional regulators present within the corynebacterial core genome. The knowledge on the physiological function of these proteins should not only contribute to our understanding of the regulation of gene expression but will also provide the basis for comprehensive modeling of transcriptional regulatory networks of these species.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

