Expression and purification of SARS coronavirus proteins using SUMO-fusions
- PMID: 15939295
- PMCID: PMC7129641
- DOI: 10.1016/j.pep.2005.02.004
Expression and purification of SARS coronavirus proteins using SUMO-fusions
Abstract
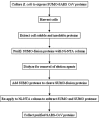
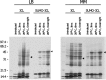
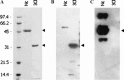
Severe acute respiratory syndrome coronavirus (SARS-CoV) proteins belong to a large group of proteins that is difficult to express in traditional expression systems. The ability to express and purify SARS-CoV proteins in large quantities is critical for basic research and for development of pharmaceutical agents. The work reported here demonstrates: (1) fusion of SUMO (small ubiquitin-related modifier), a 100 amino acid polypeptide, to the N-termini of SARS-CoV proteins dramatically enhances expression in Escherichia coli cells and (2) 6x His-tagged SUMO-fusions facilitate rapid purification of the viral proteins on a large scale. We have exploited the natural chaperoning properties of SUMO to develop an expression system suitable for proteins that cannot be expressed by traditional methodologies. A unique feature of the system is the SUMO tag, which enhances expression, facilitates purification, and can be efficiently cleaved by a SUMO-specific protease to generate native protein with a desired N-terminus. We have purified various SARS-CoV proteins under either native or denaturing conditions. These purified proteins have been used to generate highly specific polyclonal antibodies. Our study suggests that the SUMO-fusion technology will be useful for enhancing expression and purification of the viral proteins for structural and functional studies as well as for therapeutic uses.
Figures

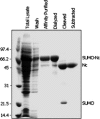

Similar articles
-
[Cloning, expression and purification of SARS coronavirus PUMC2 strain nucleocapsid protein].Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2003 Oct;25(5):504-7. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2003. PMID: 14650145 Chinese.
-
From SARS and MERS CoVs to SARS-CoV-2: Moving toward more biased codon usage in viral structural and nonstructural genes.J Med Virol. 2020 Jun;92(6):660-666. doi: 10.1002/jmv.25754. Epub 2020 Mar 16. J Med Virol. 2020. PMID: 32159237 Free PMC article.
-
[Theoretical prediction of the antigenic epitopes of severe acute respiratory syndrome (SARS-CoV) proteins and evaluation of their diagnostic value].Vopr Virusol. 2005 Sep-Oct;50(5):22-4. Vopr Virusol. 2005. PMID: 16250594 Russian.
-
An overall picture of SARS coronavirus (SARS-CoV) genome-encoded major proteins: structures, functions and drug development.Curr Pharm Des. 2006;12(35):4539-53. doi: 10.2174/138161206779010459. Curr Pharm Des. 2006. PMID: 17168760 Review.
-
Recent developments in the virology and antiviral research of severe acute respiratory syndrome coronavirus.Infect Disord Drug Targets. 2007 Mar;7(1):29-41. doi: 10.2174/187152607780090739. Infect Disord Drug Targets. 2007. PMID: 17346209 Review.
Cited by
-
Rapid Biophysical Characterization and NMR Spectroscopy Structural Analysis of Small Proteins from Bacteria and Archaea.Chembiochem. 2020 Apr 17;21(8):1178-1187. doi: 10.1002/cbic.201900677. Epub 2020 Jan 21. Chembiochem. 2020. PMID: 31705614 Free PMC article.
-
Efficient and rapid protein expression and purification of small high disulfide containing sweet protein brazzein in E. coli.Protein Expr Purif. 2008 Apr;58(2):263-8. doi: 10.1016/j.pep.2007.11.009. Epub 2007 Dec 3. Protein Expr Purif. 2008. PMID: 18221889 Free PMC article.
-
Zinc mesoporphyrin induces rapid and marked degradation of the transcription factor Bach1 and up-regulates HO-1.Biochim Biophys Acta. 2008 Mar;1779(3):195-203. doi: 10.1016/j.bbagrm.2008.01.006. Epub 2008 Feb 14. Biochim Biophys Acta. 2008. PMID: 18325350 Free PMC article.
-
SUMOylation in α-Synuclein Homeostasis and Pathology.Front Aging Neurosci. 2020 Jun 25;12:167. doi: 10.3389/fnagi.2020.00167. eCollection 2020. Front Aging Neurosci. 2020. PMID: 32670048 Free PMC article. Review.
-
Comparison of SUMO fusion technology with traditional gene fusion systems: enhanced expression and solubility with SUMO.Protein Sci. 2006 Jan;15(1):182-9. doi: 10.1110/ps.051812706. Epub 2005 Dec 1. Protein Sci. 2006. PMID: 16322573 Free PMC article.
References
-
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. - PubMed
-
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. - PubMed
-
- Snijder E., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr L.L., Poon Y., Guan Y., Rozanov M., Spaan W.J., Gorbalenya A.E. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331:991–1004. - PMC - PubMed
-
- McIntosh K. Coronaviruses: a comparative review. Curr. Top. Microbiol. Immunol. 1974;63:85–129.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

