A general method for greatly improving the affinity of antibodies by using combinatorial libraries
- PMID: 15939870
- PMCID: PMC1143585
- DOI: 10.1073/pnas.0503543102
A general method for greatly improving the affinity of antibodies by using combinatorial libraries
Abstract
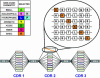
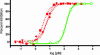
Look-through mutagenesis (LTM) is a multidimensional mutagenesis method that simultaneously assesses and optimizes combinatorial mutations of selected amino acids. The process focuses on a precise distribution within one or more complementarity determining region (CDR) domains and explores the synergistic contribution of amino acid side-chain chemistry. LTM was applied to an anti-TNF-alpha antibody, D2E7, which is a challenging test case, because D2E7 was highly optimized (K(d) = 1 nM) by others. We selected and incorporated nine amino acids, representative of the major chemical functionalities, individually at every position in each CDR and across all six CDRs (57 aa). Synthetic oligonucleotides, each introducing one amino acid mutation throughout the six CDRs, were pooled to generate segregated libraries containing single mutations in one, two, and/or three CDRs for each V(H) and V(L) domain. Corresponding antibody libraries were displayed on the cell surface of yeast. After positive binding selection, 38 substitutions in 21 CDR positions were identified that resulted in higher affinity binding to TNF-alpha. These beneficial mutations in both V(H) and V(L) were represented in two combinatorial beneficial mutagenesis libraries and selected by FACS to produce a convergence of variants that exhibit between 500- and 870-fold higher affinities. Importantly, these enhanced affinities translate to a 15- to 30-fold improvement in in vitro TNF-alpha neutralization in an L929 bioassay. Thus, this LTM/combinatorial beneficial mutagenesis strategy generates a comprehensive energetic map of the antibody-binding site in a facile and rapid manner and should be broadly applicable to the affinity maturation of antibodies and other proteins.
Figures


Similar articles
-
Isolation of picomolar affinity anti-c-erbB-2 single-chain Fv by molecular evolution of the complementarity determining regions in the center of the antibody binding site.J Mol Biol. 1996 Nov 8;263(4):551-67. doi: 10.1006/jmbi.1996.0598. J Mol Biol. 1996. PMID: 8918938
-
Humanization of a murine monoclonal antibody by simultaneous optimization of framework and CDR residues.J Mol Biol. 1999 Nov 19;294(1):151-62. doi: 10.1006/jmbi.1999.3141. J Mol Biol. 1999. PMID: 10556035
-
Affinity maturation of a humanized rat antibody for anti-RAGE therapy: comprehensive mutagenesis reveals a high level of mutational plasticity both inside and outside the complementarity-determining regions.J Mol Biol. 2009 May 8;388(3):541-58. doi: 10.1016/j.jmb.2009.03.019. Epub 2009 Mar 13. J Mol Biol. 2009. PMID: 19285987
-
Selection and evolution of high-affinity human anti-viral antibodies.Trends Biotechnol. 1996 Jul;14(7):230-4. doi: 10.1016/0167-7799(96)10029-9. Trends Biotechnol. 1996. PMID: 8771795 Review.
-
Latest technologies for the enhancement of antibody affinity.Adv Drug Deliv Rev. 2006 Aug 7;58(5-6):657-70. doi: 10.1016/j.addr.2006.01.025. Epub 2006 May 22. Adv Drug Deliv Rev. 2006. PMID: 16828920 Review.
Cited by
-
An in silico investigation on the binding site preference of PD-1 and PD-L1 for designing antibodies for targeted cancer therapy.PLoS One. 2024 Jul 25;19(7):e0304270. doi: 10.1371/journal.pone.0304270. eCollection 2024. PLoS One. 2024. PMID: 39052609 Free PMC article.
-
Simultaneous affinity maturation and developability enhancement using natural liability-free CDRs.MAbs. 2022 Jan-Dec;14(1):2115200. doi: 10.1080/19420862.2022.2115200. MAbs. 2022. PMID: 36068722 Free PMC article.
-
An affinity-enhanced neutralizing antibody against the membrane-proximal external region of human immunodeficiency virus type 1 gp41 recognizes an epitope between those of 2F5 and 4E10.J Virol. 2007 Apr;81(8):4033-43. doi: 10.1128/JVI.02588-06. Epub 2007 Feb 7. J Virol. 2007. PMID: 17287272 Free PMC article.
-
Combining deep mutational scanning to heatmap of HLA class II binding of immunogenic sequences to preserve functionality and mitigate predicted immunogenicity.Front Immunol. 2023 Jul 28;14:1197919. doi: 10.3389/fimmu.2023.1197919. eCollection 2023. Front Immunol. 2023. PMID: 37575221 Free PMC article.
-
Identification of human single-chain antibodies with broad reactivity for noroviruses.Protein Eng Des Sel. 2014 Oct;27(10):339-49. doi: 10.1093/protein/gzu023. Epub 2014 Jun 19. Protein Eng Des Sel. 2014. PMID: 24946948 Free PMC article.
References
-
- Huse, W. D., Sastry, L., Iverson, S. A., Kang, A. S., Alting-Mees, M., Burton, D. R., Benkovic, S. J. & Lerner, R. A. (1989) Science 246 1275-1281. - PubMed
-
- McCafferty, J., Griffiths, A. D., Winter, G. & Chiswell, D. J. (1990) Nature 348 552-554. - PubMed
-
- Low, N. M., Holliger, P. H. & Winter, G. (1996) J. Mol. Biol. 260 359-368. - PubMed
-
- Nishimiya, Y., Tsumoto, K., Shiroishi, M., Yutani, K. & Kumagai, I. (2000) J. Biol. Chem. 275 12813-128120. - PubMed
-
- Yang, W. P., Green, K., Pinz-Sweeney, S., Briones, A. T., Burton, D. R. & Barbas, C. F., 3rd (1995) J. Mol. Biol. 254 392-403. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

