Characterization of two avian MHC-like genes reveals an ancient origin of the CD1 family
- PMID: 15939884
- PMCID: PMC1150814
- DOI: 10.1073/pnas.0500105102
Characterization of two avian MHC-like genes reveals an ancient origin of the CD1 family
Abstract
Many of the genes that comprise the vertebrate adaptive immune system are conserved across wide evolutionary time scales. Most notably, homologs of the mammalian MHC gene family have been found in virtually all jawed vertebrates, including sharks, bony fishes, reptiles, and birds. The CD1 family of antigen-presenting molecules are related to the MHC class I family but have evolved to bind and present lipid antigens to T cells. Here, we describe two highly divergent nonclassical MHC class I genes found in the chicken (Gallus gallus) that have sequence homology to the mammalian CD1 family of proteins. One of the chicken CD1 genes expresses a full-length transcript, whereas the other has multiple splice variants. Both Southern blot and single nucleotide polymorphism analysis indicates that chicken CD1 is relatively nonpolymorphic. Moreover, cross-hybridizing bands are present in other bird species, suggesting broad conservation in the avian class. Northern analysis of chicken tissue shows a high level of CD1 expression in the bursa and spleen. In addition, molecular modeling predicts that the potential antigen-binding pocket is probably hydrophobic, a universal characteristic of CD1 molecules. Genomic analysis indicates that the CD1 genes are located on chicken chromosome 16 and maps to within 200 kb of the chicken MHC B locus, suggesting that CD1 genes diverged from classical MHC genes while still linked to the major histocompatibility complex locus. The existence of CD1 genes in an avian species suggests that the origin of CD1 extends deep into the evolutionary history of terrestrial vertebrates.
Figures

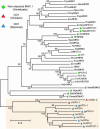
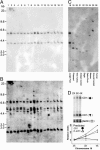

Comment in
-
Bird genes give new insights into the origins of lipid antigen presentation.Proc Natl Acad Sci U S A. 2005 Jun 14;102(24):8399-400. doi: 10.1073/pnas.0503313102. Epub 2005 Jun 6. Proc Natl Acad Sci U S A. 2005. PMID: 15939875 Free PMC article. No abstract available.
References
-
- Flajnik, M. F. & Kasahara, M. (2001) Immunity 15, 351-362. - PubMed
-
- Calabi, F. & Milstein, C. (1986) Nature 323, 540-543. - PubMed
-
- Dascher, C. C. & Brenner, M. B. (2003) Trends Immunol. 24, 412-418. - PubMed
-
- Brigl, M. & Brenner, M. B. (2004) Annu. Rev. Immunol. 22, 817-890. - PubMed
-
- Vincent, M. S., Gumperz, J. E. & Brenner, M. B. (2003) Nat. Immunol. Rev. 4, 517-523. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

