Structural properties of promoters: similarities and differences between prokaryotes and eukaryotes
- PMID: 15939933
- PMCID: PMC1143579
- DOI: 10.1093/nar/gki627
Structural properties of promoters: similarities and differences between prokaryotes and eukaryotes
Abstract
During the process of transcription, RNA polymerase can exactly locate a promoter sequence in the complex maze of a genome. Several experimental studies and computational analyses have shown that the promoter sequences apparently possess some special properties, such as unusual DNA structures and low stability, which make them distinct from the rest of the genome. But most of these studies have been carried out on a particular set of promoter sequences or on promoter sequences from similar organisms. To examine whether the promoters from a wide variety of organisms share these special properties, we have carried out an analysis of sets of promoters from bacteria, vertebrates and plants. These promoters were analyzed with respect to the prediction of three different properties, such as DNA curvature, bendability and stability, which are relevant to transcription. All the promoter sequences are predicted to share certain features, such as stability and bendability profiles, but there are significant differences in DNA curvature profiles and nucleotide composition between the different organisms. These similarities and differences are correlated with some of the known facts about transcription process in the promoters from the three groups of organisms.
Figures
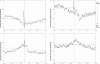
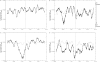
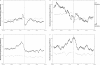


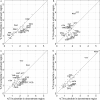
Similar articles
-
Evolutionary trends of GC/AT distribution patterns in promoters.Mol Phylogenet Evol. 2011 Aug;60(2):228-35. doi: 10.1016/j.ympev.2011.04.015. Epub 2011 May 7. Mol Phylogenet Evol. 2011. PMID: 21554969
-
DNA structure in human RNA polymerase II promoters.J Mol Biol. 1998 Aug 28;281(4):663-73. doi: 10.1006/jmbi.1998.1972. J Mol Biol. 1998. PMID: 9710538
-
Curved DNA in promoter sequences.In Silico Biol. 1999-2000;1(4):183-96. In Silico Biol. 1999. PMID: 11479933
-
Role of DNA sequence based structural features of promoters in transcription initiation and gene expression.Curr Opin Struct Biol. 2014 Apr;25:77-85. doi: 10.1016/j.sbi.2014.01.007. Epub 2014 Feb 4. Curr Opin Struct Biol. 2014. PMID: 24503515 Review.
-
Similarities and dissimilarities between plant UsnRNAs and their equivalents in other eukaryotes with respect to structure, complexing with proteins, gene organization and function. A review.Acta Biochim Biophys Hung. 1990;25(1-2):67-85. Acta Biochim Biophys Hung. 1990. PMID: 2151839 Review. No abstract available.
Cited by
-
Genome-Wide Prediction of Transcription Start Sites in Conifers.Int J Mol Sci. 2022 Feb 3;23(3):1735. doi: 10.3390/ijms23031735. Int J Mol Sci. 2022. PMID: 35163661 Free PMC article.
-
ProSOM: core promoter prediction based on unsupervised clustering of DNA physical profiles.Bioinformatics. 2008 Jul 1;24(13):i24-31. doi: 10.1093/bioinformatics/btn172. Bioinformatics. 2008. PMID: 18586720 Free PMC article.
-
Synthetic promoter design for new microbial chassis.Biochem Soc Trans. 2016 Jun 15;44(3):731-7. doi: 10.1042/BST20160042. Biochem Soc Trans. 2016. PMID: 27284035 Free PMC article. Review.
-
DNA sequence encodes the position of DNA supercoils.Elife. 2018 Dec 7;7:e36557. doi: 10.7554/eLife.36557. Elife. 2018. PMID: 30523779 Free PMC article.
-
Ambiguity in logic-based models of gene regulatory networks: An integrative multi-perturbation analysis.PLoS One. 2018 Nov 20;13(11):e0206976. doi: 10.1371/journal.pone.0206976. eCollection 2018. PLoS One. 2018. PMID: 30458000 Free PMC article.
References
-
- Bucher P. Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J. Mol. Biol. 1990;212:563–578. - PubMed
-
- Buckle M., Buc H. Fine mapping of DNA single-stranded regions using base-specific chemical probes: study of an open complex formed between RNA polymerase and the lac UV5 promoter. Biochemistry. 1989;28:4388–4396. - PubMed
-
- Chen Y.F., Helmann J.D. DNA-melting at the Bacillus subtilis flagellin promoter nucleates near −10 and expands unidirectionally. J. Mol. Biol. 1997;267:47–59. - PubMed
-
- Craig M.L., Suh W.C., Record M.T., Jr HO. and DNase I probing of E sigma 70 RNA polymerase–lambda PR promoter open complexes: Mg2+ binding and its structural consequences at the transcription start site. Biochemistry. 1995;34:15624–15632. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

