Nuclear distribution and chromatin association of DNA polymerase alpha-primase is affected by TEV protease cleavage of Cdc23 (Mcm10) in fission yeast
- PMID: 15941470
- PMCID: PMC1182370
- DOI: 10.1186/1471-2199-6-13
Nuclear distribution and chromatin association of DNA polymerase alpha-primase is affected by TEV protease cleavage of Cdc23 (Mcm10) in fission yeast
Abstract
Background: Cdc23/Mcm10 is required for the initiation and elongation steps of DNA replication but its biochemical function is unclear. Here, we probe its function using a novel approach in fission yeast, involving Cdc23 cleavage by the TEV protease.
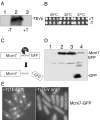
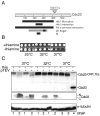
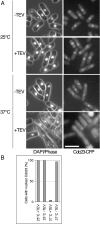
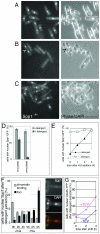
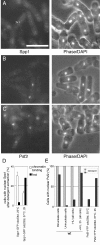
Results: Insertion of a TEV protease cleavage site into Cdc23 allows in vivo removal of the C-terminal 170 aa of the protein by TEV protease induction, resulting in an S phase arrest. This C-terminal fragment of Cdc23 is not retained in the nucleus after cleavage, showing that it lacks a nuclear localization signal and ability to bind to chromatin. Using an in situ chromatin binding procedure we have determined how the S phase chromatin association of DNA polymerase alpha-primase and the GINS (Sld5-Psf1-Psf2-Psf3) complex is affected by Cdc23 inactivation. The chromatin binding and sub-nuclear distribution of DNA primase catalytic subunit (Spp1) is affected by Cdc23 cleavage and also by inactivation of Cdc23 using a degron allele, implying that DNA polymerase alpha-primase function is dependent on Cdc23. In contrast to the effect on Spp1, the chromatin association of the Psf2 subunit of the GINS complex is not affected by Cdc23 inactivation.
Conclusion: An important function of Cdc23 in the elongation step of DNA replication may be to assist in the docking of DNA polymerase alpha-primase to chromatin.
Figures

Similar articles
-
Fission yeast Cdc23/Mcm10 functions after pre-replicative complex formation to promote Cdc45 chromatin binding.Mol Biol Cell. 2003 Sep;14(9):3876-87. doi: 10.1091/mbc.e03-02-0090. Epub 2003 Jun 13. Mol Biol Cell. 2003. PMID: 12972571 Free PMC article.
-
GINS inactivation phenotypes reveal two pathways for chromatin association of replicative alpha and epsilon DNA polymerases in fission yeast.Mol Biol Cell. 2009 Feb;20(4):1213-22. doi: 10.1091/mbc.e08-04-0429. Epub 2008 Dec 24. Mol Biol Cell. 2009. PMID: 19109429 Free PMC article.
-
The essential schizosaccharomyces pombe cdc23 DNA replication gene shares structural and functional homology with the Saccharomyces cerevisiae DNA43 (MCM10) gene.Curr Genet. 1998 Sep;34(3):164-71. doi: 10.1007/s002940050382. Curr Genet. 1998. PMID: 9745018
-
RNA insertion in DNA as the imprint moiety: the fission yeast paradigm.Curr Genet. 2019 Dec;65(6):1301-1306. doi: 10.1007/s00294-019-00991-x. Epub 2019 May 10. Curr Genet. 2019. PMID: 31076844 Review.
-
Origins and complexes: the initiation of DNA replication.J Exp Bot. 2001 Feb;52(355):193-202. J Exp Bot. 2001. PMID: 11283163 Review.
Cited by
-
Cloning, expression, purification, crystallization and preliminary X-ray diffraction analysis of the central zinc-binding domain of the human Mcm10 DNA-replication factor.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2008 Jun 1;64(Pt 6):495-7. doi: 10.1107/S1744309108011640. Epub 2008 May 16. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2008. PMID: 18540058 Free PMC article.
-
Human Mcm10 regulates the catalytic subunit of DNA polymerase-alpha and prevents DNA damage during replication.Mol Biol Cell. 2007 Oct;18(10):4085-95. doi: 10.1091/mbc.e06-12-1148. Epub 2007 Aug 15. Mol Biol Cell. 2007. PMID: 17699597 Free PMC article.
-
Domain architecture and biochemical characterization of vertebrate Mcm10.J Biol Chem. 2008 Feb 8;283(6):3338-3348. doi: 10.1074/jbc.M706267200. Epub 2007 Dec 6. J Biol Chem. 2008. PMID: 18065420 Free PMC article.
-
Physical interactions between Mcm10, DNA, and DNA polymerase alpha.J Biol Chem. 2009 Sep 4;284(36):24662-72. doi: 10.1074/jbc.M109.020438. Epub 2009 Jul 16. J Biol Chem. 2009. PMID: 19608746 Free PMC article.
-
Cdt1 proteolysis is promoted by dual PIP degrons and is modulated by PCNA ubiquitylation.Nucleic Acids Res. 2011 Aug;39(14):5978-90. doi: 10.1093/nar/gkr222. Epub 2011 Apr 14. Nucleic Acids Res. 2011. PMID: 21493688 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

