Global and Hox-specific roles for the MLL1 methyltransferase
- PMID: 15941828
- PMCID: PMC1150839
- DOI: 10.1073/pnas.0503072102
Global and Hox-specific roles for the MLL1 methyltransferase
Abstract
The mixed-lineage leukemia (MLL1/ALL-1/HRX) histone methyltransferase is involved in the epigenetic maintenance of transcriptional memory and the pathogenesis of human leukemias. To understand its role in cell type specification, we determined the human genomic binding sites of MLL1. We found that MLL1 functions as a human equivalent of yeast Set1. Like Set1, MLL1 localizes with RNA polymerase II (Pol II) to the 5' end of actively transcribed genes, where histone H3 lysine 4 trimethylation occurs. Consistent with this global role in transcription, MLL1 also localizes to microRNA (miRNA) loci that are involved in leukemia and hematopoiesis. In contrast to the 5' proximal binding behavior at most protein-coding genes, MLL1 occupies an extensive domain within a transcriptionally active region of the HoxA cluster. The ability of MLL1 to serve as a start site-specific global transcriptional regulator and to participate in larger chromatin domains at the Hox genes reveals dual roles for MLL1 in maintenance of cellular identity.
Figures

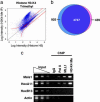
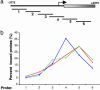


References
-
- Fischle, W., Wang, Y. & Allis, C. D. (2003) Curr. Opin. Cell Biol. 15, 172-183. - PubMed
-
- Sims, R. J., III, Nishioka, K. & Reinberg, D. (2003) Trends Genet. 19, 629-639. - PubMed
-
- Santos-Rosa, H., Schneider, R., Bannister, A. J., Sherriff, J., Bernstein, B. E., Emre, N. C., Schreiber, S. L., Mellor, J. & Kouzarides, T. (2002) Nature 419, 407-411. - PubMed
-
- Ng, H. H., Robert, F., Young, R. A. & Struhl, K. (2003) Mol. Cell 11, 709-719. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

