Differential in vivo binding dynamics of somatic and oocyte-specific linker histones in oocytes and during ES cell nuclear transfer
- PMID: 15944219
- PMCID: PMC1182324
- DOI: 10.1091/mbc.e05-04-0350
Differential in vivo binding dynamics of somatic and oocyte-specific linker histones in oocytes and during ES cell nuclear transfer
Abstract
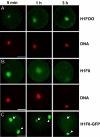
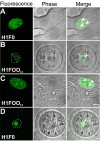
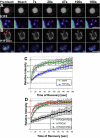
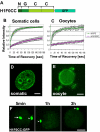
The embryonic genome is formed by fusion of a maternal and a paternal genome. To accommodate the resulting diploid genome in the fertilized oocyte dramatic global genome reorganizations must occur. The higher order structure of chromatin in vivo is critically dependent on architectural chromatin proteins, with the family of linker histone proteins among the most critical structural determinants. Although somatic cells contain numerous linker histone variants, only one, H1FOO, is present in mouse oocytes. Upon fertilization H1FOO rapidly populates the introduced paternal genome and replaces sperm-specific histone-like proteins. The same dynamic replacement occurs upon introduction of a nucleus during somatic cell nuclear transfer. To understand the molecular basis of this dynamic histone replacement process, we compared the localization and binding dynamics of somatic H1 and oocyte-specific H1FOO and identified the molecular determinants of binding to either oocyte or somatic chromatin in living cells. We find that although both histones associate readily with chromatin in nuclei of somatic cells, only H1FOO is capable of correct chromatin association in the germinal vesicle stage oocyte nuclei. This specificity is generated by the N-terminal and globular domains of H1FOO. Measurement of in vivo binding properties of the H1 variants suggest that H1FOO binds chromatin more tightly than somatic linker histones. We provide evidence that both the binding properties of linker histones as well as additional, active processes contribute to the replacement of somatic histones with H1FOO during nuclear transfer. These results provide the first mechanistic insights into the crucial step of linker histone replacement as it occurs during fertilization and somatic cell nuclear transfer.
Figures


Similar articles
-
Rapid replacement of somatic linker histones with the oocyte-specific linker histone H1foo in nuclear transfer.Dev Biol. 2004 Feb 1;266(1):76-86. doi: 10.1016/j.ydbio.2003.10.004. Dev Biol. 2004. PMID: 14729479
-
Oocyte-specific linker histone H1foo interacts with Esrrb to induce chromatin decondensation at specific gene loci.Biochem Biophys Res Commun. 2021 Jul 5;561:165-171. doi: 10.1016/j.bbrc.2021.05.033. Epub 2021 May 21. Biochem Biophys Res Commun. 2021. PMID: 34023782
-
DNA hypomethylation circuit of the mouse oocyte-specific histone H1foo gene in female germ cell lineage.Biol Reprod. 2008 May;78(5):816-21. doi: 10.1095/biolreprod.107.066522. Epub 2008 Jan 9. Biol Reprod. 2008. PMID: 18184919
-
Linker histone transitions during mammalian oogenesis and embryogenesis.Dev Genet. 1998;22(1):17-30. doi: 10.1002/(SICI)1520-6408(1998)22:1<17::AID-DVG3>3.0.CO;2-A. Dev Genet. 1998. PMID: 9499577 Review.
-
H3.3 replacement facilitates epigenetic reprogramming of donor nuclei in somatic cell nuclear transfer embryos.Nucleus. 2014 Sep-Oct;5(5):369-75. doi: 10.4161/nucl.36231. Nucleus. 2014. PMID: 25482190 Review.
Cited by
-
Maternal control of early embryogenesis in mammals.Reprod Fertil Dev. 2015 Jul;27(6):880-96. doi: 10.1071/RD14441. Reprod Fertil Dev. 2015. PMID: 25695370 Free PMC article. Review.
-
Oocyte-type linker histone B4 is required for transdifferentiation of somatic cells in vivo.FASEB J. 2010 Sep;24(9):3462-7. doi: 10.1096/fj.10-159285. Epub 2010 May 11. FASEB J. 2010. PMID: 20460584 Free PMC article.
-
H1foo Has a Pivotal Role in Qualifying Induced Pluripotent Stem Cells.Stem Cell Reports. 2016 Jun 14;6(6):825-833. doi: 10.1016/j.stemcr.2016.04.015. Epub 2016 May 26. Stem Cell Reports. 2016. PMID: 27237376 Free PMC article.
-
Characterization of somatic cell nuclear reprogramming by oocytes in which a linker histone is required for pluripotency gene reactivation.Proc Natl Acad Sci U S A. 2010 Mar 23;107(12):5483-8. doi: 10.1073/pnas.1000599107. Epub 2010 Mar 8. Proc Natl Acad Sci U S A. 2010. PMID: 20212135 Free PMC article.
-
EGFP-tagged core and linker histones diffuse via distinct mechanisms within living cells.Biophys J. 2006 Sep 15;91(6):2326-36. doi: 10.1529/biophysj.105.079343. Epub 2006 Jun 30. Biophys J. 2006. PMID: 16815908 Free PMC article.
References
-
- Bustin, M., Catez, F., and Lim, J. H. (2005). The dynamics of histone h1 function in chromatin. Mol. Cell 17, 617-620. - PubMed
-
- Churikov, D., Zalenskaya, I. A., and Zalensky, A. O. (2004). Male germline-specific histones in mouse and man. Cytogenet. Genome Res. 105, 203-214. - PubMed
-
- Draves, P. H., Lowary, P. T., and Widom, J. (1992). Co-operative binding of the globular domain of histone H5 to DNA. J. Mol. Biol. 225, 1105-1121. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

