Large-scale profiling of Rab GTPase trafficking networks: the membrome
- PMID: 15944222
- PMCID: PMC1182321
- DOI: 10.1091/mbc.e05-01-0062
Large-scale profiling of Rab GTPase trafficking networks: the membrome
Abstract
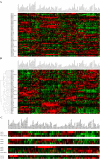
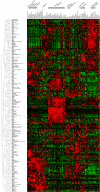
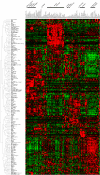
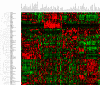
Rab GTPases and SNARE fusion proteins direct cargo trafficking through the exocytic and endocytic pathways of eukaryotic cells. We have used steady state mRNA expression profiling and computational hierarchical clustering methods to generate a global overview of the distribution of Rabs, SNAREs, and coat machinery components, as well as their respective adaptors, effectors, and regulators in 79 human and 61 mouse nonredundant tissues. We now show that this systems biology approach can be used to define building blocks for membrane trafficking based on Rab-centric protein activity hubs. These Rab-regulated hubs provide a framework for an integrated coding system, the membrome network, which regulates the dynamics of the specialized membrane architecture of differentiated cells. The distribution of Rab-regulated hubs illustrates a number of facets that guides the overall organization of subcellular compartments of cells and tissues through the activity of dynamic protein interaction networks. An interactive website for exploring datasets comprising components of the Rab-regulated hubs that define the membrome of different cell and organ systems in both human and mouse is available at http://www.membrome.org/.
Figures


Similar articles
-
Exploring trafficking GTPase function by mRNA expression profiling: use of the SymAtlas web-application and the Membrome datasets.Methods Enzymol. 2005;403:1-10. doi: 10.1016/S0076-6879(05)03001-6. Methods Enzymol. 2005. PMID: 16473572
-
An evolutionary perspective on eukaryotic membrane trafficking.Adv Exp Med Biol. 2007;607:73-83. doi: 10.1007/978-0-387-74021-8_6. Adv Exp Med Biol. 2007. PMID: 17977460 Review.
-
Rab GTPases: master regulators that establish the secretory and endocytic pathways.Mol Biol Cell. 2017 Mar 15;28(6):712-715. doi: 10.1091/mbc.E16-10-0737. Mol Biol Cell. 2017. PMID: 28292916 Free PMC article. Review.
-
Rab proteins: the key regulators of intracellular vesicle transport.Exp Cell Res. 2014 Oct 15;328(1):1-19. doi: 10.1016/j.yexcr.2014.07.027. Epub 2014 Aug 1. Exp Cell Res. 2014. PMID: 25088255 Review.
-
Rab-mediated vesicle trafficking in cancer.J Biomed Sci. 2016 Oct 6;23(1):70. doi: 10.1186/s12929-016-0287-7. J Biomed Sci. 2016. PMID: 27716280 Free PMC article. Review.
Cited by
-
Phylogenetic reconstruction and evolution of the Rab GTPase gene family in Amoebozoa.Small GTPases. 2022 Jan;13(1):100-113. doi: 10.1080/21541248.2021.1903794. Epub 2021 Mar 29. Small GTPases. 2022. PMID: 33779495 Free PMC article.
-
PhosphoregDB: the tissue and sub-cellular distribution of mammalian protein kinases and phosphatases.BMC Bioinformatics. 2006 Feb 20;7:82. doi: 10.1186/1471-2105-7-82. BMC Bioinformatics. 2006. PMID: 16504016 Free PMC article.
-
Maturation of the mammalian secretome.Genome Biol. 2007;8(4):211. doi: 10.1186/gb-2007-8-4-211. Genome Biol. 2007. PMID: 17472737 Free PMC article. Review.
-
The Hsp90 chaperone complex regulates GDI-dependent Rab recycling.Mol Biol Cell. 2006 Aug;17(8):3494-507. doi: 10.1091/mbc.e05-12-1096. Epub 2006 May 10. Mol Biol Cell. 2006. PMID: 16687576 Free PMC article.
-
Post-transcriptional regulation of myotube elongation and myogenesis by Hoi Polloi.Development. 2013 Sep;140(17):3645-56. doi: 10.1242/dev.095596. Development. 2013. PMID: 23942517 Free PMC article.
References
-
- Abdul-Ghani, M., Gougeon, P. Y., Prosser, D. C., Da-Silva, L. F., and Ngsee, J. K. (2001). PRA isoforms are targeted to distinct membrane compartments. J. Biol. Chem. 276, 6225-6233. - PubMed
-
- Allan, B. B., Moyer, B. D., and Balch, W. E. (2000). Rab1 recruitment of p115 into a cis-SNARE complex: programming budding COPII vesicles for fusion. Science 289, 444-448. - PubMed
-
- Antonny, B., and Schekman, R. (2001). ER export: public transportation by the COPII coach. Curr. Opin. Cell Biol. 13, 438-443. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

