Basal body duplication and maintenance require one member of the Tetrahymena thermophila centrin gene family
- PMID: 15944224
- PMCID: PMC1182301
- DOI: 10.1091/mbc.e04-10-0919
Basal body duplication and maintenance require one member of the Tetrahymena thermophila centrin gene family
Abstract
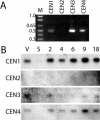
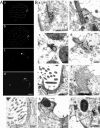
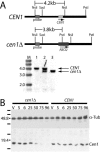
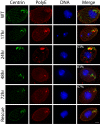
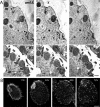
Centrins, small calcium binding EF-hand proteins, function in the duplication of a variety of microtubule organizing centers. These include centrioles in humans, basal bodies in green algae, and spindle pole bodies in yeast. The ciliate Tetrahymena thermophila contains at least four centrin genes as determined by sequence homology, and these have distinct localization and expression patterns. CEN1's role at the basal body was examined more closely. The Cen1 protein localizes primarily to two locations: one is the site at the base of the basal body where duplication is initiated. The other is the transition zone between the basal body and axoneme. CEN1 is an essential gene, the deletion of which results in the loss of basal bodies, which is likely due to defects in both basal body duplication and basal body maintenance. Analysis of the three other centrins indicates that two of them function at microtubule-rich structures unique to ciliates, whereas the fourth is not expressed under conditions examined in this study, although when artificially expressed it localizes to basal bodies. This study provides evidence that in addition to its previously known function in the duplication of basal bodies, centrin is also important for the integrity of these organelles.
Figures

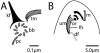
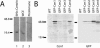
Similar articles
-
The two human centrin homologues have similar but distinct functions at Tetrahymena basal bodies.Mol Biol Cell. 2012 Dec;23(24):4766-77. doi: 10.1091/mbc.E12-06-0454. Epub 2012 Oct 19. Mol Biol Cell. 2012. PMID: 23087207 Free PMC article.
-
The two domains of centrin have distinct basal body functions in Tetrahymena.Mol Biol Cell. 2011 Jul 1;22(13):2221-34. doi: 10.1091/mbc.E11-02-0151. Epub 2011 May 11. Mol Biol Cell. 2011. PMID: 21562224 Free PMC article.
-
Sfr1, a Tetrahymena thermophila Sfi1 Repeat Protein, Modulates the Production of Cortical Row Basal Bodies.mSphere. 2016 Nov 16;1(6):e00257-16. doi: 10.1128/mSphere.00257-16. eCollection 2016 Nov-Dec. mSphere. 2016. PMID: 27904881 Free PMC article.
-
A mechanistic view on the evolutionary origin for centrin-based control of centriole duplication.J Cell Physiol. 2007 Nov;213(2):420-8. doi: 10.1002/jcp.21226. J Cell Physiol. 2007. PMID: 17694534 Review.
-
Tetrahymena basal bodies.Cilia. 2016 Jan 19;5:1. doi: 10.1186/s13630-016-0022-8. eCollection 2015. Cilia. 2016. PMID: 26793300 Free PMC article. Review.
Cited by
-
DisAp-dependent striated fiber elongation is required to organize ciliary arrays.J Cell Biol. 2014 Dec 22;207(6):705-15. doi: 10.1083/jcb.201409123. J Cell Biol. 2014. PMID: 25533842 Free PMC article.
-
The two SAS-6 homologs in Tetrahymena thermophila have distinct functions in basal body assembly.Mol Biol Cell. 2009 Mar;20(6):1865-77. doi: 10.1091/mbc.e08-08-0838. Epub 2009 Jan 21. Mol Biol Cell. 2009. PMID: 19158390 Free PMC article.
-
Multiciliated cells.Curr Biol. 2014 Oct 6;24(19):R973-82. doi: 10.1016/j.cub.2014.08.047. Curr Biol. 2014. PMID: 25291643 Free PMC article. Review.
-
New Tetrahymena basal body protein components identify basal body domain structure.J Cell Biol. 2007 Sep 10;178(6):905-12. doi: 10.1083/jcb.200703109. Epub 2007 Sep 4. J Cell Biol. 2007. PMID: 17785518 Free PMC article.
-
Differential localization and functional specialization of centrin analogs in the parasitic ciliate Trichodina pediculus.Protoplasma. 2016 Sep;253(5):1385-8. doi: 10.1007/s00709-015-0878-2. Epub 2015 Sep 4. Protoplasma. 2016. PMID: 26340903
References
-
- Adams, I. R., and Kilmartin, J. V. (2000). Spindle pole body duplication: a model for centrosome duplication? Trends Cell Biol. 10, 329–335. - PubMed
-
- Alberts, B., Johnson, A., Lewis, J., Raff, M., Roberts, K., and Walter, P. (2002). Molecular Biology of the Cell, New York: Garland Science.
-
- Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smoth, J. A., and Struhl, K. (1997). Current Protocols in Molecular Biology, New York: John Wiley & Sons, Inc.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

