Selection for recombination in structured populations
- PMID: 15944358
- PMCID: PMC1456186
- DOI: 10.1534/genetics.104.039982
Selection for recombination in structured populations
Abstract
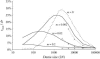
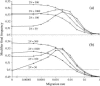
In finite populations, linkage disequilibria generated by the interaction of drift and directional selection (Hill-Robertson effect) can select for sex and recombination, even in the absence of epistasis. Previous models of this process predict very little advantage to recombination in large panmictic populations. In this article we demonstrate that substantial levels of linkage disequilibria can accumulate by drift in the presence of selection in populations of any size, provided that the population is subdivided. We quantify (i) the linkage disequilibrium produced by the interaction of drift and selection during the selective sweep of beneficial alleles at two loci in a subdivided population and (ii) the selection for recombination generated by these disequilibria. We show that, in a population subdivided into n demes of large size N, both the disequilibrium and the selection for recombination are equivalent to that expected in a single population of a size intermediate between the size of each deme (N) and the total size (nN), depending on the rate of migration among demes, m. We also show by simulations that, with small demes, the selection for recombination is stronger than both that expected in an unstructured population (m = 1 - 1/n) and that expected in a set of isolated demes (m = 0). Indeed, migration maintains polymorphisms that would otherwise be lost rapidly from small demes, while population structure maintains enough local stochasticity to generate linkage disequilibria. These effects are also strong enough to overcome the twofold cost of sex under strong selection when sex is initially rare. Overall, our results show that the stochastic theories of the evolution of sex apply to a much broader range of conditions than previously expected.
Figures





References
-
- Barton, N. H., 1995. a A general model for the evolution of recombination. Genet. Res. 65: 123–144. - PubMed
-
- Barton, N. H., and K. S. Gale, 1993. Genetic analysis of hybrid zones, pp. 13–45 in Hybrid Zones and the Evolutionary Process, edited by R. G. Harrison. Oxford University Press, Oxford.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

