Polygenic mutation in Drosophila melanogaster: Mapping spontaneous mutations affecting sensory bristle number
- PMID: 15944368
- PMCID: PMC1449762
- DOI: 10.1534/genetics.104.032581
Polygenic mutation in Drosophila melanogaster: Mapping spontaneous mutations affecting sensory bristle number
Abstract
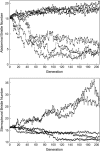
Our ability to predict long-term responses to artificial and natural selection, and understand the mechanisms by which naturally occurring variation for quantitative traits is maintained, depends on detailed knowledge of the properties of spontaneous polygenic mutations, including the quantitative trait loci (QTL) at which mutations occur, mutation rates, and mutational effects. These parameters can be estimated by mapping QTL that cause divergence between mutation-accumulation lines that have been established from an inbred base population and selected for high and low trait values. Here, we have utilized quantitative complementation to deficiencies to map QTL at which spontaneous mutations affecting Drosophila abdominal and sternopleural bristle number have occurred in 11 replicate lines during 206 generations of divergent selection. Estimates of the numbers of mutations were consistent with diploid per-character mutation rates for bristle traits of 0.03. The ratio of the per-character mutation rate to total mutation rate (0.023) implies that >2% of the genome could affect just one bristle trait and that there must be extensive pleiotropy for quantitative phenotypes. The estimated mutational effects were not, however, additive and exhibited dependency on genetic background consistent with diminishing epistasis. However, these inferences must be tempered by the potential for epistatic interactions between spontaneous mutations and QTL affecting bristle number on the deficiency-bearing chromosomes, which could lead to overestimates in numbers of QTL and inaccurate inference of gene action.
Figures
References
-
- Adams, M. D., S. Celniker, R. A. Holt, C. A. Evans, J. D. Gocayne et al., 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185–2195. - PubMed
-
- Barton, N., and M. Turelli, 1989. Evolutionary quantitative genetics: How little do we know? Annu. Rev. Genet. 23: 337–370. - PubMed
-
- Clayton, G. A., and A. Robertson, 1955. Mutation and quantitative variation. Am. Nat. 89: 151–158.
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases