A complementation method for functional analysis of mammalian genes
- PMID: 15944448
- PMCID: PMC1145195
- DOI: 10.1093/nar/gni093
A complementation method for functional analysis of mammalian genes
Abstract
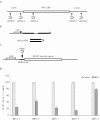
Our progress in understanding mammalian gene function has lagged behind that of gene identification. New methods for mammalian gene functional analysis are needed to accelerate the process. In yeast, the powerful genetic shuffle system allows deletion of any chromosomal gene by homologous recombination and episomal expression of a mutant allele in the same cell. Here, we report a method for mammalian cells, which employs a helper-dependent adenoviral (HD-Ad) vector to synthesize small hairpin (sh) RNAs to knock-down the expression of an endogenous gene by targeting untranslated regions (UTRs). The vector simultaneously expresses an exogenous version of the same gene (wild-type or mutant allele) lacking the UTRs for functional analysis. We demonstrated the utility of the method by using PRPF3, which encodes the human RNA splicing factor Hprp3p. Recently, missense mutations in PRPF3 were found to cause autosomal-dominant Retinitis Pigmentosa, a form of genetic eye diseases affecting the retina. We knocked-down endogenous PRPF3 in multiple cell lines and rescued the phenotype (cell death) with exogenous PRPF3 cDNA, thereby creating a genetic complementation method. Because Ad vectors can efficiently transduce a wide variety of cell types, and many tissues in vivo, this method could have a wide application for gene function studies.
Figures





References
-
- Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. - PubMed
-
- Venter J.C., Adams M.D., Myers E.W., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., et al. The sequence of the human genome. Science. 2001;291:1304–1351. - PubMed
-
- Shoemaker D.D., Schadt E.E., Armour C.D., He Y.D., Garrett-Engele P., McDonagh P.D., Loerch P.M., Leonardson A., Lum P.Y., Cavet G., et al. Experimental annotation of the human genome using microarray technology. Nature. 2001;409:922–927. - PubMed
-
- Shi Y. Mammalian RNAi for the masses. Trends Genet. 2003;19:9–12. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

