Human Rad51 filaments on double- and single-stranded DNA: correlating regular and irregular forms with recombination function
- PMID: 15944450
- PMCID: PMC1145190
- DOI: 10.1093/nar/gki640
Human Rad51 filaments on double- and single-stranded DNA: correlating regular and irregular forms with recombination function
Abstract
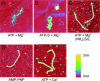
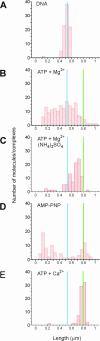
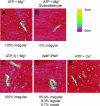
Recombinase proteins assembled into helical filaments on DNA are believed to be the catalytic core of homologous recombination. The assembly, disassembly and dynamic rearrangements of this structure must drive the DNA strand exchange reactions of homologous recombination. The sensitivity of eukaryotic recombinase activity to reaction conditions in vitro suggests that the status of bound nucleotide cofactors is important for function and possibly for filament structure. We analyzed nucleoprotein filaments formed by the human recombinase Rad51 in a variety of conditions on double-stranded and single-stranded DNA by scanning force microscopy. Regular filaments with extended double-stranded DNA correlated with active in vitro recombination, possibly due to stabilizing the DNA products of these assays. Though filaments formed readily on single-stranded DNA, they were very rarely regular structures. The irregular structure of filaments on single-stranded DNA suggests that Rad51 monomers are dynamic in filaments and that regular filaments are transient. Indeed, single molecule force spectroscopy of Rad51 filament assembly and disassembly in magnetic tweezers revealed protein association and disassociation from many points along the DNA, with kinetics different from those of RecA. The dynamic rearrangements of proteins and DNA within Rad51 nucleoprotein filaments could be key events driving strand exchange in homologous recombination.
Figures
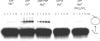

Similar articles
-
Real-time assembly and disassembly of human RAD51 filaments on individual DNA molecules.Nucleic Acids Res. 2007;35(17):5646-57. doi: 10.1093/nar/gkm629. Epub 2007 Aug 20. Nucleic Acids Res. 2007. PMID: 17709342 Free PMC article.
-
Visualizing the assembly of human Rad51 filaments on double-stranded DNA.J Mol Biol. 2006 Oct 27;363(3):713-28. doi: 10.1016/j.jmb.2006.08.046. Epub 2006 Aug 22. J Mol Biol. 2006. PMID: 16979659
-
Homologous genetic recombination as an intrinsic dynamic property of a DNA structure induced by RecA/Rad51-family proteins: a possible advantage of DNA over RNA as genomic material.Proc Natl Acad Sci U S A. 2001 Jul 17;98(15):8425-32. doi: 10.1073/pnas.111005198. Proc Natl Acad Sci U S A. 2001. PMID: 11459985 Free PMC article. Review.
-
Stimulation by Rad52 of yeast Rad51-mediated recombination.Nature. 1998 Jan 22;391(6665):404-7. doi: 10.1038/34943. Nature. 1998. PMID: 9450759
-
Regulation of DNA strand exchange in homologous recombination.DNA Repair (Amst). 2010 Dec 10;9(12):1264-72. doi: 10.1016/j.dnarep.2010.09.014. DNA Repair (Amst). 2010. PMID: 20971042 Review.
Cited by
-
Biophysical characterization of DNA binding from single molecule force measurements.Phys Life Rev. 2010 Sep;7(3):299-341. doi: 10.1016/j.plrev.2010.06.001. Epub 2010 Jun 4. Phys Life Rev. 2010. PMID: 20576476 Free PMC article. Review.
-
Swi5-Sfr1 stimulates Rad51 recombinase filament assembly by modulating Rad51 dissociation.Proc Natl Acad Sci U S A. 2018 Oct 23;115(43):E10059-E10068. doi: 10.1073/pnas.1812753115. Epub 2018 Oct 8. Proc Natl Acad Sci U S A. 2018. PMID: 30297419 Free PMC article.
-
Contributions of the RAD51 N-terminal domain to BRCA2-RAD51 interaction.Nucleic Acids Res. 2013 Oct;41(19):9020-32. doi: 10.1093/nar/gkt691. Epub 2013 Aug 8. Nucleic Acids Res. 2013. PMID: 23935068 Free PMC article.
-
Two distinct conformational states define the interaction of human RAD51-ATP with single-stranded DNA.EMBO J. 2018 Apr 3;37(7):e98162. doi: 10.15252/embj.201798162. Epub 2018 Mar 5. EMBO J. 2018. PMID: 29507080 Free PMC article.
-
Chromatin dynamics during repair of chromosomal DNA double-strand breaks.Epigenomics. 2009 Dec;1(2):371-85. doi: 10.2217/epi.09.22. Epigenomics. 2009. PMID: 20495614 Free PMC article. Review.
References
-
- DiCapua E., Engel A., Stasiak A., Kooller T. Characterization of complexes between recA protein and duplex DNA by electron microscopy. J. Mol. Biol. 1982;157:87–103. - PubMed
-
- Dunn K., Chrysogelos S., Griffith J. Electron microscopic visualization of RecA-DNA filaments: evidence for cyclic extension of duplex DNA. Cell. 1982;28:757–765. - PubMed
-
- Flory J., Radding C.M. Visualization of RecA protein and its association with DNA: a priming effect of single-strand binding protein. Cell. 1982;28:747–756. - PubMed
-
- Yu X., VanLoock M.S., Yang S., Reese J.T., Egelman E.H. What is the structure of the RecA-DNA filament? Curr. Protein Pept. Sci. 2004;5:73–79. - PubMed
-
- Liu Y., Stasiak A.Z., Masson J.Y., McIlwraith M.J., Stasiak A., West S.C. Conformational changes modulate the activity of human RAD51 protein. J. Mol. Biol. 2004;337:817–827. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

