Metabolic engineering in the -omics era: elucidating and modulating regulatory networks
- PMID: 15944454
- PMCID: PMC1197421
- DOI: 10.1128/MMBR.69.2.197-216.2005
Metabolic engineering in the -omics era: elucidating and modulating regulatory networks
Abstract
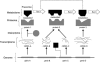
The importance of regulatory control in metabolic processes is widely acknowledged, and several enquiries (both local and global) are being made in understanding regulation at various levels of the metabolic hierarchy. The wealth of biological information has enabled identifying the individual components (genes, proteins, and metabolites) of a biological system, and we are now in a position to understand the interactions between these components. Since phenotype is the net result of these interactions, it is immensely important to elucidate them not only for an integrated understanding of physiology, but also for practical applications of using biological systems as cell factories. We present some of the recent "-omics" approaches that have expanded our understanding of regulation at the gene, protein, and metabolite level, followed by analysis of the impact of this progress on the advancement of metabolic engineering. Although this review is by no means exhaustive, we attempt to convey our ideology that combining global information from various levels of metabolic hierarchy is absolutely essential in understanding and subsequently predicting the relationship between changes in gene expression and the resulting phenotype. The ultimate aim of this review is to provide metabolic engineers with an overview of recent advances in complementary aspects of regulation at the gene, protein, and metabolite level and those involved in fundamental research with potential hurdles in the path to implementing their discoveries in practical applications.
Figures
References
-
- Aebersold, R., and M. Mann. 2003. Mass spectrometry-based proteomics. Nature 422:198-207. - PubMed
-
- Agarwal, A. K., P. D. Rogers, S. R. Baerson, M. R. Jacob, K. S. Barker, J. D. Cleary, L. A. Walker, D. G. Nagle, and A. M. Clark. 2003. Genome-wide expression profiling of the response to polyene, pyrimidine, azole, and echinocandin antifungal agents in Saccharomyces cerevisiae. J. Biol. Chem. 278:34998-35015. - PubMed
-
- Anderson, L., and J. Seilhamer. 1997. A comparison of selected mRNA and protein abundances in human liver. Electrophoresis 18:533-537. - PubMed
-
- Aravin, A. A., M. Lagos-Quintana, A. Yalcin, M. Zavolan, D. Marks, B. Snyder, T. Gaasterland, J. Meyer, and T. Tuschl. 2003. The small RNA profile during Drosophila melanogaster development. Dev. Cell 5:337-350. - PubMed
-
- Arfin, S. M., A. D. Long, E. T. Ito, L. Tolleri, M. M. Riehle, E. S. Paegle, and G. W. Hatfield. 2000. Global gene expression profiling in Escherichia coli K12. The effects of integration host factor. J. Biol. Chem. 275:29672-29684. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

