Zn2+-induced reversible dissociation of subunit Rpn10/p54 of the Drosophila 26 S proteasome
- PMID: 15946124
- PMCID: PMC1276928
- DOI: 10.1042/BJ20050523
Zn2+-induced reversible dissociation of subunit Rpn10/p54 of the Drosophila 26 S proteasome
Abstract
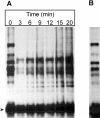
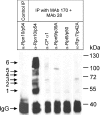
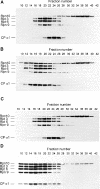
In the presence of Zn2+, the Drosophila 26 S proteasome disassembles into RP (regulatory particle) and CP (catalytic particle), this process being accompanied by the dissociation of subunit Rpn10/p54, the ubiquitin receptor subunit of the proteasome. The dissociation of Rpn10/p54 induces extensive rearrangements within the lid subcomplex of the RP, while the structure of the ATPase ring of the base subcomplex seems to be maintained. As a consequence of the dissociation of the RP, the peptidase activity of the 26 S proteasome is lost. The Zn2+-induced structural and functional changes are fully reversible; removal of Zn2+ is followed by reassociation of subunit Rpn10/p54 to the RP, reassembly of the 26 S proteasome and resumption of the peptidase activity. After the Zn2+-induced dissociation, Rpn10/p54 interacts with a set of non-proteasomal proteins. Hsp82 (heat-shock protein 82) has been identified by MS as the main Rpn10/p54-interacting protein, suggesting its role in the reassembly of the 26 S proteasome after Zn2+ removal. The physiological relevance of another Rpn10/p54-interacting protein, the Smt3 SUMO (small ubiquitin-related modifier-1)-activating enzyme, detected by chemical cross-linking, has been confirmed by yeast two-hybrid analysis. Besides the Smt3 SUMO-activating enzyme, the Ubc9 SUMO-conjugating enzyme also exhibited in vivo interaction with the 5'-half of Rpn10/p54 in yeast cells. The mechanism of 26 S proteasome disassembly after ATP depletion is clearly different from that induced by Zn2+. Rpn10/p54 is permanently RP-bound during the ATP-dependent assembly-disassembly cycle, but during the Zn2+ cycle it reversibly shuttles between the RP-bound and free states.
Figures





Similar articles
-
Developmental-stage-specific regulation of the polyubiquitin receptors in Drosophila melanogaster.J Cell Sci. 2009 Sep 1;122(Pt 17):3083-92. doi: 10.1242/jcs.049049. Epub 2009 Aug 4. J Cell Sci. 2009. PMID: 19654212
-
Deletion of proteasomal subunit S5a/Rpn10/p54 causes lethality, multiple mitotic defects and overexpression of proteasomal genes in Drosophila melanogaster.J Cell Sci. 2003 Mar 15;116(Pt 6):1023-33. doi: 10.1242/jcs.00332. J Cell Sci. 2003. PMID: 12584246
-
¹H, ¹³C and ¹⁵N resonance assignments of the VWA domain of Saccharomyces cerevisiae Rpn10, a regulatory subunit of 26S proteasome.Biomol NMR Assign. 2014 Oct;8(2):391-4. doi: 10.1007/s12104-013-9525-z. Epub 2013 Sep 14. Biomol NMR Assign. 2014. PMID: 24037519
-
Assembly manual for the proteasome regulatory particle: the first draft.Biochem Soc Trans. 2010 Feb;38(Pt 1):6-13. doi: 10.1042/BST0380006. Biochem Soc Trans. 2010. PMID: 20074027 Free PMC article. Review.
-
Chaperone-driven proteasome assembly.Biochem Soc Trans. 2008 Oct;36(Pt 5):807-12. doi: 10.1042/BST0360807. Biochem Soc Trans. 2008. PMID: 18793141 Review.
Cited by
-
E2-25K SUMOylation inhibits proteasome for cell death during cerebral ischemia/reperfusion.Cell Death Dis. 2016 Dec 29;7(12):e2573. doi: 10.1038/cddis.2016.428. Cell Death Dis. 2016. PMID: 28032866 Free PMC article.
-
Killer proteases and little strokes--how the things that do not kill you make you stronger.J Cereb Blood Flow Metab. 2007 Apr;27(4):655-68. doi: 10.1038/sj.jcbfm.9600380. Epub 2006 Aug 9. J Cereb Blood Flow Metab. 2007. PMID: 16896349 Free PMC article. Review.
-
Molecular characterization of the Rpt1/p48B ATPase subunit of the Drosophila melanogaster 26S proteasome.Mol Genet Genomics. 2007 Jul;278(1):17-29. doi: 10.1007/s00438-007-0223-3. Epub 2007 Apr 12. Mol Genet Genomics. 2007. PMID: 17429695
-
Coordination chemistry suggests that independently observed benefits of metformin and Zn2+ against COVID-19 are not independent.Biometals. 2024 Aug;37(4):983-1022. doi: 10.1007/s10534-024-00590-5. Epub 2024 Apr 5. Biometals. 2024. PMID: 38578560 Free PMC article.
-
Insights into the molecular architecture of the 26S proteasome.Proc Natl Acad Sci U S A. 2009 Jul 21;106(29):11943-7. doi: 10.1073/pnas.0905081106. Epub 2009 Jul 6. Proc Natl Acad Sci U S A. 2009. PMID: 19581588 Free PMC article.
References
-
- Pickart C. M. Back to the future with ubiquitin. Cell (Cambridge, Mass.) 2004;116:181–190. - PubMed
-
- Groll M., Hubert R. Substrate access and processing by the 20S proteasome core particle. Int. J. Biochem. Cell Biol. 2003;35:606–616. - PubMed
-
- Wenzel T., Baumeister W. Conformational constrains in protein degradation by the 20S proteasome. Nat. Struct. Biol. 1995;2:199–204. - PubMed
-
- Löwe J., Stock D., Jap B., Zwickl P., Baumeister W., Huber R. Crystal structure of the 20S proteasome from the archeon T. acidophilum at 3.4 Å resolution. Science. 1995;268:533–539. - PubMed
-
- Groll M., Ditzel L., Löwe J., Stock D., Bochtler M., Bartunik H. D., Huber R. Structure of the 20SA proteasome from yeast at 2.4 Å resolution. Nature (London) 1997;386:463–471. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

