Inferring yeast cell cycle regulators and interactions using transcription factor activities
- PMID: 15949038
- PMCID: PMC1180827
- DOI: 10.1186/1471-2164-6-90
Inferring yeast cell cycle regulators and interactions using transcription factor activities
Abstract
Background: Since transcription factors are often regulated at the post-transcriptional level, their activities, rather than expression levels may provide valuable information for investigating functions and their interactions. The recently developed Network Component Analysis (NCA) and its generalized form (gNCA) provide a robust framework for deducing the transcription factor activities (TFAs) from various types of DNA microarray data and transcription factor-gene connectivity. The goal of this work is to demonstrate the utility of TFAs in inferring transcription factor functions and interactions in Saccharomyces cerevisiae cell cycle regulation.
Results: Using gNCA, we determined 74 TFAs from both wild type and fkh1 fkh2 deletion mutant microarray data encompassing 1529 ORFs. We hypothesized that transcription factors participating in the cell cycle regulation exhibit cyclic activity profiles. This hypothesis was supported by the TFA profiles of known cell cycle factors and was used as a basis to uncover other potential cell cycle factors. By combining the results from both cluster analysis and periodicity analysis, we recovered nearly 90% of the known cell cycle regulators, and identified 5 putative cell cycle-related transcription factors (Dal81, Hap2, Hir2, Mss11, and Rlm1). In addition, by analyzing expression data from transcription factor knockout strains, we determined 3 verified (Ace2, Ndd1, and Swi5) and 4 putative interaction partners (Cha4, Hap2, Fhl1, and Rts2) of the forkhead transcription factors. Sensitivity of TFAs to connectivity errors was determined to provide confidence level of these predictions.
Conclusion: By subjecting TFA profiles to analyses based upon physiological signatures we were able to identify cell cycle related transcription factors consistent with current literature, transcription factors with potential cell cycle dependent roles, and interactions between transcription factors.
Figures
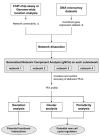
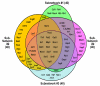
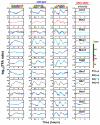




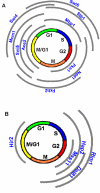
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

