Effects of organelle shape on fluorescence recovery after photobleaching
- PMID: 15951382
- PMCID: PMC1366654
- DOI: 10.1529/biophysj.104.057885
Effects of organelle shape on fluorescence recovery after photobleaching
Abstract
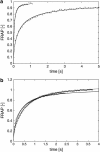
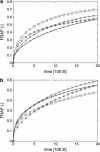
The determination of diffusion coefficients from fluorescence recovery data is often complicated by geometric constraints imposed by the complex shapes of intracellular compartments. To address this issue, diffusion of proteins in the lumen of the endoplasmic reticulum (ER) is studied using cell biological and computational methods. Fluorescence recovery after photobleaching (FRAP) experiments are performed in tissue culture cells expressing GFP-KDEL, a soluble, fluorescent protein, in the ER lumen. The three-dimensional (3D) shape of the ER is determined by confocal microscopy and computationally reconstructed. Within these ER geometries diffusion of solutes is simulated using the method of particle strength exchange. The simulations are compared to experimental FRAP curves of GFP-KDEL in the same ER region. Comparisons of simulations in the 3D ER shapes to simulations in open 3D space show that the constraints imposed by the spatial confinement result in two- to fourfold underestimation of the molecular diffusion constant in the ER if the geometry is not taken into account. Using the same molecular diffusion constant in different simulations, the observed speed of fluorescence recovery varies by a factor of 2.5, depending on the particular ER geometry and the location of the bleached area. Organelle shape considerably influences diffusive transport and must be taken into account when relating experimental photobleaching data to molecular diffusion coefficients. This novel methodology combines experimental FRAP curves with high accuracy computer simulations of diffusion in the same ER geometry to determine the molecular diffusion constant of the solute in the particular ER lumen.
Figures


References
-
- Lippincott-Schwartz, J., E. Snapp, and A. Kenworthy. 2001. Studying protein dynamics in living cells. Nat. Rev. Mol. Cell Biol. 2:444–456. - PubMed
-
- Phair, R. D., and T. Misteli. 2000. High mobility of proteins in the mammalian cell nucleus. Nature. 404:604–609. - PubMed
-
- Nehls, S., E. L. Snapp, N. B. Cole, K. J. M. Zaal, A. K. Kenworthy, T. H. Roberts, J. Ellenberg, J. F. Presley, E. Siggia, and J. Lippincott-Schwartz. 2000. Dynamics and retention of misfolded proteins in native ER membranes. Nat. Cell Biol. 2:288–295. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

